
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,106,935 – 4,107,169 |
| Length | 234 |
| Max. P | 0.999847 |

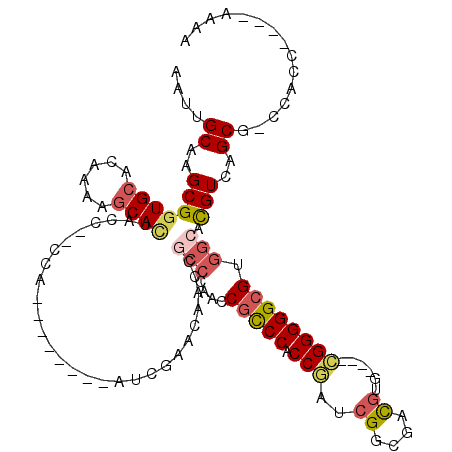
| Location | 4,106,935 – 4,107,035 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.97 |
| Mean single sequence MFE | -30.84 |
| Consensus MFE | -19.92 |
| Energy contribution | -20.84 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4106935 100 - 22224390 AAUUGCAAGCGGUGCACAAAACCACUAC---CCA--------AUCGCCCAAUCGCCCAACCGCCCACCGAUCGGCGACGUG----CGGGGGCGUGGCACGUCAGCG-CCACC----AAAA .((((..((.(((.......))).))..---.))--------)).((((...(((.(...((((........))))..).)----)).))))(((((.(....).)-)))).----.... ( -31.40) >DroSec_CAF1 118740 103 - 1 AAUUGCAAGCGGUGCACAAAAGCAUUACCCCCCA--------AUCGAACAACCGCCCAACCGCCCACCGAUCGGCGACGUG----CGGGGGCGUGGCACGUCAGCG-CCACC----AAAA ........(((((((......)).........(.--------...)....)))))......((((.(((..((....))..----)))))))(((((.(....).)-)))).----.... ( -32.80) >DroSim_CAF1 119909 112 - 1 AAUUGCAAGCGGUGCACAAAAGCAUUACCCCCCAAUCGCCCAAUCGAACAACCGCCCAACCGCCCACCGAUCGGCGACGUG----CGGGGGCGUGGCACGUCAGCGACCGCC----AAAA ........(((((((....................(((......)))......(((....(((((.(((..((....))..----)))))))).)))......)).))))).----.... ( -34.60) >DroEre_CAF1 125090 84 - 1 AAUUGCAAGCGGUGCACAAAAGCACUACC---------------------------CAACCGCCCACCAAUCGGCGACGUG----CGGGGGCGUGGCACGUCAGCG-CCACC----AAAA ..........(((((......)))))...---------------------------.....((((.((...((....))..----.))))))(((((.(....).)-)))).----.... ( -29.70) >DroAna_CAF1 148245 107 - 1 AGUUGCAAGCGCUGCACAA-AGCACCAC---UCU--------AACACACAACCACCUCCACAUCCGCCUCUCGUGUGUAUGUUAAAGGGGGCGUGGCAUGUCAGCA-CCACCACCGAAAA .((((((.(((((......-))).....---...--------....(((..((.(((..((((.((((....).))).))))...)))))..))))).)).)))).-............. ( -25.70) >consensus AAUUGCAAGCGGUGCACAAAAGCACUACC__CCA________AUCGAACAACCGCCCAACCGCCCACCGAUCGGCGACGUG____CGGGGGCGUGGCACGUCAGCG_CCACC____AAAA ....((..(((((((......))))............................(((....(((((.(((..((....))......)))))))).))).)))..))............... (-19.92 = -20.84 + 0.92)


| Location | 4,106,966 – 4,107,075 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.51 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -14.63 |
| Energy contribution | -15.05 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4106966 109 - 22224390 UAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCACAAAAUUGCAAGCGGUGCACAAAACCACUAC---CCA--------AUCGCCCAAUCGCCCAACCGCCCACCGAUCGGCGACGU ...........((((.......))))....(((((((......))).((.(((.......))).))..---...--------...))))..(((((....((.....))...)))))... ( -24.90) >DroSec_CAF1 118771 112 - 1 UAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGAGCACAAAAUUGCAAGCGGUGCACAAAAGCAUUACCCCCCA--------AUCGAACAACCGCCCAACCGCCCACCGAUCGGCGACGU ..((.......((((.......))))....(((.(((......)))..(..((((......))))..)..))).--------...)).............((((........)))).... ( -21.30) >DroSim_CAF1 119941 120 - 1 UAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCACAAAAUUGCAAGCGGUGCACAAAAGCAUUACCCCCCAAUCGCCCAAUCGAACAACCGCCCAACCGCCCACCGAUCGGCGACGU .(((.......((((.......))))....(((((((......)))..(((((((......))............(((......)))...)))))......))))...)))((....)). ( -26.30) >DroEre_CAF1 125121 93 - 1 UAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCUCAAAAUUGCAAGCGGUGCACAAAAGCACUACC---------------------------CAACCGCCCACCAAUCGGCGACGU ...........((((.......))))....((((((........))....(((((......)))))...---------------------------.....))))......((....)). ( -21.60) >DroYak_CAF1 118415 92 - 1 UAUCUAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCACAAAAUUGCAAGCGGUGCACAAAAGCACUACC---------------------------CAA-CGCCCACCACCCGGCGACGU ...........((((.......))))....(((((((......)))....(((((......)))))...---------------------------...-.))))......((....)). ( -22.30) >DroAna_CAF1 148284 106 - 1 UAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGCGA--AAAGUUGCAAGCGCUGCACAA-AGCACCAC---UCU--------AACACACAACCACCUCCACAUCCGCCUCUCGUGUGUAU ...........((((.......))))....((((((--....))))....(((......-))).))..---...--------.((((((.......................)))))).. ( -16.50) >consensus UAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCACAAAAUUGCAAGCGGUGCACAAAAGCACUACC__CCA________AUCG_ACAACCGCCCAACCGCCCACCGAUCGGCGACGU ...........((((.......))))....((((((........))....(((((......)))))...................................))))......((....)). (-14.63 = -15.05 + 0.42)


| Location | 4,107,035 – 4,107,155 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -39.18 |
| Consensus MFE | -36.14 |
| Energy contribution | -36.78 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4107035 120 + 22224390 UUGUGCGCCCUAAUGCAAUUUUGUUUUGCUUAAUUUGAUAGGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCACACCAGCAACA .(((..((.....((((((((((((((((((...(((.((((((((((((((((......))).)))))))))......)))).)))))))))....)))))))))))).....)).))) ( -39.60) >DroSec_CAF1 118843 120 + 1 UUGUGCUCCCUAAUGCAAUUUUGUUUUGCUUAAUUUGAUAGGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCACACCAGCAACA ...((((......((((((((((((((((((...(((.((((((((((((((((......))).)))))))))......)))).)))))))))....))))))))))))....))))... ( -40.00) >DroSim_CAF1 120021 120 + 1 UUGUGCGCCCUAAUGCAAUUUUGUUUUGCUUAAUUUGAUAGGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCACACCAGCAACA .(((..((.....((((((((((((((((((...(((.((((((((((((((((......))).)))))))))......)))).)))))))))....)))))))))))).....)).))) ( -39.60) >DroEre_CAF1 125174 120 + 1 UUGAGCGCCCUAAUGCAAUUUUGUUUUGCUUAAUUUGAUAGGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCAAACCAGCAACA ......((.....((((((((((((((((((...(((.((((((((((((((((......))).)))))))))......)))).)))))))))....)))))))))))).....)).... ( -39.00) >DroYak_CAF1 118467 120 + 1 UUGUGCGCCCUAAUGCAAUUUUGUUUUGCUUAAUUAGAUAGGCGUUGUCAGCGGAGCUGCCAGCCUGACAAUGUGUCUACCUAACAGGAGCAACAAUAGCAAAAUGGCACACCAGCAACA .(((((((......)).((((((((((((((..((((.((((((((((((((((.....)).).))))))))..))))).))))...))))))....)))))))).)))))......... ( -37.70) >consensus UUGUGCGCCCUAAUGCAAUUUUGUUUUGCUUAAUUUGAUAGGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCACACCAGCAACA ...(((.......((((((((((((((((((....((.((((((((((((((((......))).)))))))))......)))).)).))))))....)))))))))))).....)))... (-36.14 = -36.78 + 0.64)



| Location | 4,107,035 – 4,107,155 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -35.24 |
| Energy contribution | -36.64 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4107035 120 - 22224390 UGUUGCUGGUGUGCAAUUUUGCCAUUGUUGCUCCUGUUAGGUGGACACAUUGUCAGGCUGGCAGCUCAGCUGACAACGCCUAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCACAA (((((((...(((((((((((...(((((.....((.((((((......(((((((.((((....))))))))))))))))).)).....)))))..)))))))))))...)))).))). ( -40.90) >DroSec_CAF1 118843 120 - 1 UGUUGCUGGUGUGCAAUUUUGCCAUUGUUGCUCCUGUUAGGUGGACACAUUGUCAGGCUGGCAGCUCAGCUGACAACGCCUAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGAGCACAA ((((.((...(((((((((((...(((((.....((.((((((......(((((((.((((....))))))))))))))))).)).....)))))..)))))))))))..)).))))... ( -41.00) >DroSim_CAF1 120021 120 - 1 UGUUGCUGGUGUGCAAUUUUGCCAUUGUUGCUCCUGUUAGGUGGACACAUUGUCAGGCUGGCAGCUCAGCUGACAACGCCUAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCACAA (((((((...(((((((((((...(((((.....((.((((((......(((((((.((((....))))))))))))))))).)).....)))))..)))))))))))...)))).))). ( -40.90) >DroEre_CAF1 125174 120 - 1 UGUUGCUGGUUUGCAAUUUUGCCAUUGUUGCUCCUGUUAGGUGGACACAUUGUCAGGCUGGCAGCUCAGCUGACAACGCCUAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCUCAA ....(((....((((((((((...(((((.....((.((((((......(((((((.((((....))))))))))))))))).)).....)))))..))))))))))....)))...... ( -37.40) >DroYak_CAF1 118467 120 - 1 UGUUGCUGGUGUGCCAUUUUGCUAUUGUUGCUCCUGUUAGGUAGACACAUUGUCAGGCUGGCAGCUCCGCUGACAACGCCUAUCUAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCACAA ........(((((((....(((.(((.(((((...(((((((((((.....))).(((((.((((...)))).))..)))))))))))..))))).....))).)))....))))))).. ( -40.90) >consensus UGUUGCUGGUGUGCAAUUUUGCCAUUGUUGCUCCUGUUAGGUGGACACAUUGUCAGGCUGGCAGCUCAGCUGACAACGCCUAUCAAAUUAAGCAAAACAAAAUUGCAUUAGGGCGCACAA (((((((...(((((((((((...(((((.....((.((((((......(((((((.((((....))))))))))))))))).)).....)))))..)))))))))))...)))).))). (-35.24 = -36.64 + 1.40)



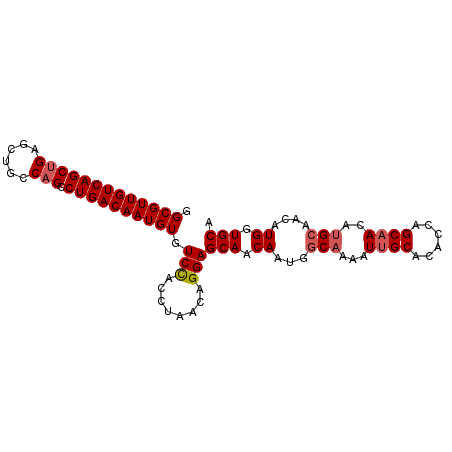
| Location | 4,107,075 – 4,107,169 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.46 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4107075 94 + 22224390 GGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCACACCAGCAACAUGCAACAUGGUGCA .(((((((((((((......))).)))))))))).(((........)))(((...((((((...((((......))))..)))..)))..))). ( -30.30) >DroSec_CAF1 118883 94 + 1 GGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCACACCAGCAACAAGAAACAUGGUGCA .(((((((((((((......))).)))))))))).(((........)))(((...(((......((((......)))).......)))..))). ( -28.42) >DroSim_CAF1 120061 94 + 1 GGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCACACCAGCAACAUGCAACAUGGUGCA .(((((((((((((......))).)))))))))).(((........)))(((...((((((...((((......))))..)))..)))..))). ( -30.30) >DroEre_CAF1 125214 94 + 1 GGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCAAACCAGCAACAUGCAACUUGGUGCA .(((((((((((((......))).)))))))))).(((........)))(((.(((..(((...((((......))))..)))...))).))). ( -31.40) >DroYak_CAF1 118507 94 + 1 GGCGUUGUCAGCGGAGCUGCCAGCCUGACAAUGUGUCUACCUAACAGGAGCAACAAUAGCAAAAUGGCACACCAGCAACAUGCAACAUGAUGCA ((.(((((((((((.....)).).))))))))(((((..((.....)).((.......)).....))))).)).(((.((((...)))).))). ( -26.60) >consensus GGCGUUGUCAGCUGAGCUGCCAGCCUGACAAUGUGUCCACCUAACAGGAGCAACAAUGGCAAAAUUGCACACCAGCAACAUGCAACAUGGUGCA .(((((((((((((......))).)))))))))).(((........)))(((.((...(((...((((......))))..)))....)).))). (-25.82 = -26.46 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:38 2006