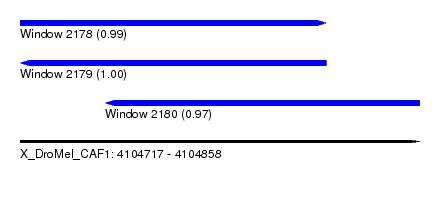
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,104,717 – 4,104,858 |
| Length | 141 |
| Max. P | 0.998980 |

| Location | 4,104,717 – 4,104,825 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.30 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -22.54 |
| Energy contribution | -22.35 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
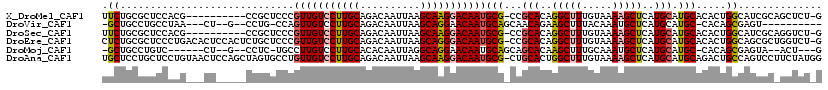
>X_DroMel_CAF1 4104717 108 + 22224390 UUCUGCGCUCCACG----------CCGCUCCCGUUGUCCUUGCAGACAAUUAAGCAAGGACAAUGCG-CCGCACAGGCUUUGUAAAAGCUCAUGCAUGCACACUGGCAUCGCAGCUCU-G ..(((((......(----------((.....((((((((((((..........)))))))))))).(-(.(((..(((((.....)))))..)))..)).....)))..)))))....-. ( -37.70) >DroVir_CAF1 189857 100 + 1 -GCUGCCUGCCUAA---CU--G--CCUG-CCAGUUGUCCUUGCAGACAAUUAAGCAGGAACAAUGCAGCAACAGAAGCUUUACAAAUGCUCAUGCAUGC-CACAGCGAGU---------- -(((((((((.(((---.(--(--.(((-(.((.....)).)))).)).))).)))))......((((((...((.((.........)))).))).)))-..))))....---------- ( -29.30) >DroSec_CAF1 116598 108 + 1 UUCUGCGCUCCACG----------CCGCUCCCGUUGUCCUUGCAGACAAUUAAGCAAGGACAAUGCG-CCGCACAGGCUUUGUAAAAGCUCAUGCAUGCACACUGGCAUCGCAGGUCU-G .((((((......(----------((.....((((((((((((..........)))))))))))).(-(.(((..(((((.....)))))..)))..)).....)))..))))))...-. ( -39.00) >DroEre_CAF1 122869 118 + 1 CUCUGCGCUCCCUGACACUCCACUCUGCUCCCGUUGUCCUUGCAGACAAUUAAGCAGGGACAAUGCG-CCGCACAGGCUUUGUAAAAGCUCAUGCAUGCACACUGGCAGCGCUGGUCU-G .((.(((((.((.(.................((((((((((((..........)))))))))))).(-(.(((..(((((.....)))))..)))..))...).)).))))).))...-. ( -38.80) >DroMoj_CAF1 176863 102 + 1 -GCUGCCUGUC------CU--G--CCUC-UGCCUUGUCCUUGCACACAAUUAGGCAGGAACAAUGCAGCAGCACAAGCUUUGCAAAUGCUCAUGCAUGC-CACAGCGAGUA--ACU---G -(((((.((((------((--(--(((.-....((((........))))..))))))).)))..)))))...((..(((.((...((((....))))..-)).)))..)).--...---. ( -32.90) >DroAna_CAF1 145509 119 + 1 UGCUCCUGCUCCUGUAACUCCAGCUAGUGCCUGUUGUCCUUGCAGACAAUUAAGCAAGGACAAUGCG-CUGCACUGGCUUUGUAAAAGCUCAUGCAUGCAGACUGCCAGUCCUUCUAUGG .((....))..........(((((.(((((..(((((((((((..........))))))))))))))-))))((((((((((((...((....)).))))))..)))))).......))) ( -41.60) >consensus UGCUGCGCUCCACG___CU_____CCGCUCCCGUUGUCCUUGCAGACAAUUAAGCAAGGACAAUGCG_CCGCACAGGCUUUGUAAAAGCUCAUGCAUGCACACUGCCAGCGC_GCUCU_G .((.............................(((((((((((..........)))))))))))(((...(((..(((((.....)))))..))).))).....)).............. (-22.54 = -22.35 + -0.19)
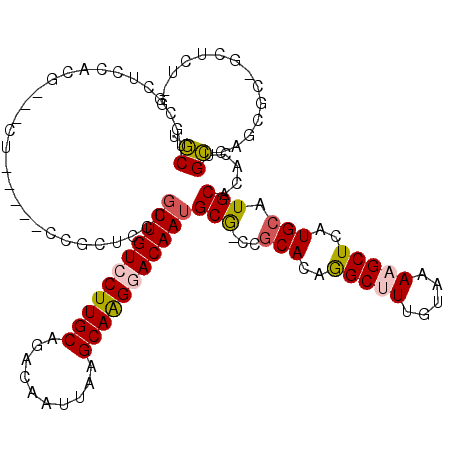
| Location | 4,104,717 – 4,104,825 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.30 |
| Mean single sequence MFE | -45.02 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.42 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4104717 108 - 22224390 C-AGAGCUGCGAUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGG-CGCAUUGUCCUUGCUUAAUUGUCUGCAAGGACAACGGGAGCGG----------CGUGGAGCGCAGAA .-....(((((((((((((((((.(((((..((((.....))))..))))))-))))))((((((((..........)))))))).........))----------)))....))))).. ( -46.90) >DroVir_CAF1 189857 100 - 1 ----------ACUCGCUGUG-GCAUGCAUGAGCAUUUGUAAAGCUUCUGUUGCUGCAUUGUUCCUGCUUAAUUGUCUGCAAGGACAACUGG-CAGG--C--AG---UUAGGCAGGCAGC- ----------......((..-(((.(((.((((.........)))).))))))..))..((((((((((((((((((((.((.....)).)-))))--)--))---))))))))).)))- ( -44.80) >DroSec_CAF1 116598 108 - 1 C-AGACCUGCGAUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGG-CGCAUUGUCCUUGCUUAAUUGUCUGCAAGGACAACGGGAGCGG----------CGUGGAGCGCAGAA .-....(((((((((((((((((.(((((..((((.....))))..))))))-))))))((((((((..........)))))))).........))----------)))....))))).. ( -47.00) >DroEre_CAF1 122869 118 - 1 C-AGACCAGCGCUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGG-CGCAUUGUCCCUGCUUAAUUGUCUGCAAGGACAACGGGAGCAGAGUGGAGUGUCAGGGAGCGCAGAG .-......(((((.(((((((((.(((((..((((.....))))..))))))-)))))).(((((((((..((((((....))))))...))))))...)))......)).))))).... ( -50.30) >DroMoj_CAF1 176863 102 - 1 C---AGU--UACUCGCUGUG-GCAUGCAUGAGCAUUUGCAAAGCUUGUGCUGCUGCAUUGUUCCUGCCUAAUUGUGUGCAAGGACAAGGCA-GAGG--C--AG------GACAGGCAGC- .---.((--(((.....)))-))..(((..(((.........)))..))).(((((.((((.(((((((..((((.(....).))))....-.)))--)--))------))))))))))- ( -41.30) >DroAna_CAF1 145509 119 - 1 CCAUAGAAGGACUGGCAGUCUGCAUGCAUGAGCUUUUACAAAGCCAGUGCAG-CGCAUUGUCCUUGCUUAAUUGUCUGCAAGGACAACAGGCACUAGCUGGAGUUACAGGAGCAGGAGCA ((......((((.....))))..........((((((..((..(((((..((-.((.((((((((((..........))))))))))...)).)).)))))..))..)))))).)).... ( -39.80) >consensus C_AGAGC_GCACUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGG_CGCAUUGUCCCUGCUUAAUUGUCUGCAAGGACAACGGGAGCGG_____AG___CAGGGAGCGCAGAA ........((((.....))))((.(((((..((((.....))))..)))))...)).((((((.(((..........))).))))))................................. (-18.86 = -19.28 + 0.42)


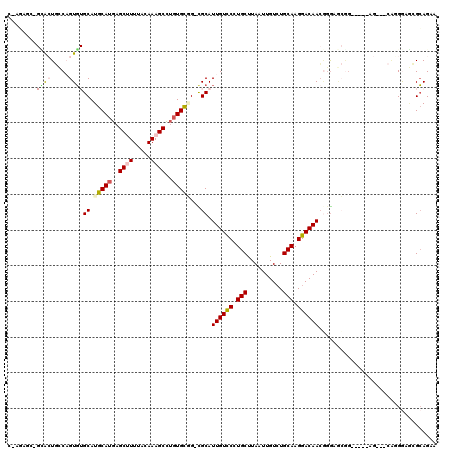
| Location | 4,104,747 – 4,104,858 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.89 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -13.09 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
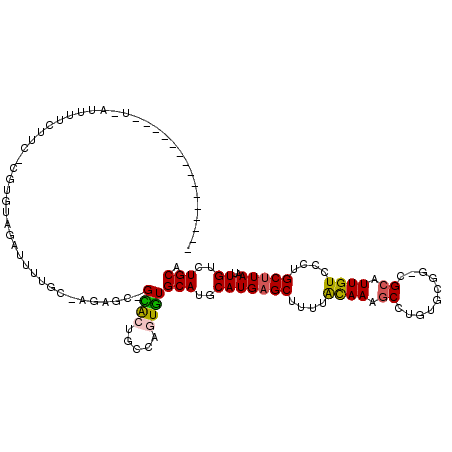
>X_DroMel_CAF1 4104747 111 - 22224390 -------AUGUGAAUUUAUUUUCUUAGCGUGUAGAUUUUGC-AGAGCUGCGAUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGG-CGCAUUGUCCUUGCUUAAUUGUCUGCA -------....((((((((...........))))))))(((-(((...((((.(.((((((((.(((((..((((.....))))..))))))-))))))).).)))).......)))))) ( -34.60) >DroGri_CAF1 153765 87 - 1 ---------------------------CAGGUGGGUGUUGCAAUAGU--UACUCGCAGUG-GCAUGCAUGAGCAUUUAUAAGACUUGUG---CUGCAUUGUUCCUGCUUAAUUGUCUGCA ---------------------------...........(((((((((--((...((((..-(((((((...((((...........)))---))))).)))..)))).))))))).)))) ( -23.00) >DroSec_CAF1 116628 104 - 1 --------------CUAAUUUUCUUCGCGUGUAGAUUUUGC-AGACCUGCGAUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGG-CGCAUUGUCCUUGCUUAAUUGUCUGCA --------------........................(((-((((..((((.(.((((((((.(((((..((((.....))))..))))))-))))))).).))))......))))))) ( -35.80) >DroEre_CAF1 122909 118 - 1 AUAUAAUAUGUGAAUCCGUUUUCUUCAUGUGUAGAUUUUGC-AGACCAGCGCUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGG-CGCAUUGUCCCUGCUUAAUUGUCUGCA .....(((((((((.........)))))))))......(((-((((.((((..(.((((((((.(((((..((((.....))))..))))))-))))))).)..)))).....))))))) ( -39.50) >DroMoj_CAF1 176891 87 - 1 ---------------------------CAGGGAACUGCUGC---AGU--UACUCGCUGUG-GCAUGCAUGAGCAUUUGCAAAGCUUGUGCUGCUGCAUUGUUCCUGCCUAAUUGUGUGCA ---------------------------..((((((.((..(---(((--.....))))..-))(((((..(((((..((...))..)))))..))))).))))))(((.......).)). ( -32.00) >DroAna_CAF1 145549 104 - 1 --------------AUAAUUUUUUUCA-GUGCAACCAUUGCCAUAGAAGGACUGGCAGUCUGCAUGCAUGAGCUUUUACAAAGCCAGUGCAG-CGCAUUGUCCUUGCUUAAUUGUCUGCA --------------...........((-((((....(((((((.........)))))))..((.(((((..((((.....))))..))))))-)))))))....(((..........))) ( -27.70) >consensus _______________U_AUUUUCUUC_CGUGUAGAUUUUGC_AGAGC_GCACUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGG_CGCAUUGUCCCUGCUUAAUUGUCUGCA ................................................((((.....))))(((..(((((((....((((.((..........)).))))....)))))..))..))). (-13.09 = -12.87 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:34 2006