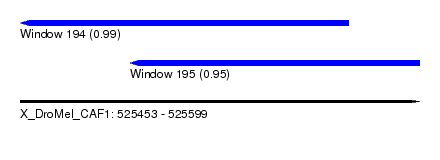
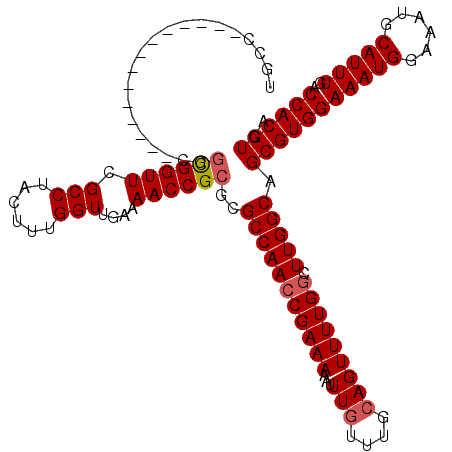
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 525,453 – 525,599 |
| Length | 146 |
| Max. P | 0.987905 |

| Location | 525,453 – 525,573 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -31.46 |
| Energy contribution | -32.06 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 525453 120 - 22224390 GAAAACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCAGAAAACGUGAAAGAAGCUAACAAGGAUAACCGGGGCAGAG .....(((((((((((((((((...(((....))))))))).))))).))))))(((((......)))))...(((((......)))))......(((.....((....))..))).... ( -34.90) >DroSec_CAF1 11938 119 - 1 GA-AACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCAGAAAACGUGAAAGAAGCUAACAAGGAUAACCGGGGCAGAG ..-..(((((((((((((((((...(((....))))))))).))))).))))))(((((......)))))...(((((......)))))......(((.....((....))..))).... ( -34.90) >DroSim_CAF1 12262 120 - 1 GAAAACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCAGAAAACGUGAAAGAAGCUAACAAGGAUAACCGGGGCAGAG .....(((((((((((((((((...(((....))))))))).))))).))))))(((((......)))))...(((((......)))))......(((.....((....))..))).... ( -34.90) >DroEre_CAF1 17022 120 - 1 GAAAACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCAGAAAACGUGAAAGAAGCUAACAAGGAUAACCGGGGCAGAG .....(((((((((((((((((...(((....))))))))).))))).))))))(((((......)))))...(((((......)))))......(((.....((....))..))).... ( -34.90) >DroYak_CAF1 10776 120 - 1 GGAAACCGCGCGCCAAGCGAAAAAUUUGUUUGGAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCAGGAAACGUGAAAGAAGCUAACAAGAAUAACCGGGGCAGAG ((...(((((((((((((..(((((((.....))))))).))))))).))))))..........((.(((...(((((......)))))...)))))............))......... ( -36.70) >consensus GAAAACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCAGAAAACGUGAAAGAAGCUAACAAGGAUAACCGGGGCAGAG .....(((((((((((((((((...(((....))))))))).))))).))))))(((((......)))))...(((((......)))))......(((.....((....))..))).... (-31.46 = -32.06 + 0.60)

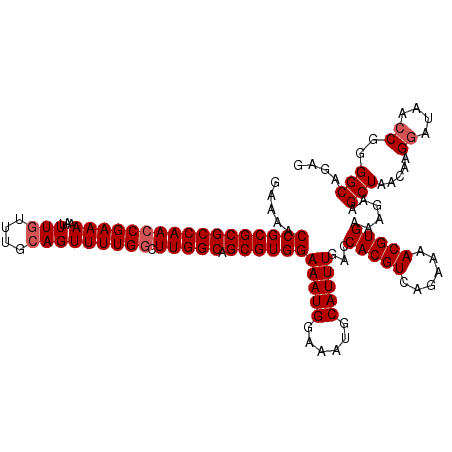
| Location | 525,493 – 525,599 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -30.82 |
| Energy contribution | -31.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 525493 106 - 22224390 UGCC--------------CGGCGGUUCGCCUACUUUGGUUGAAAACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCA ....--------------.((((...)))).....(((((.....(((((((((((((((((...(((....))))))))).))))).))))))(((((......))))))))))..... ( -35.60) >DroSec_CAF1 11978 105 - 1 UGCC--------------CGGCGGUUCGCCUACUUUGGUUGA-AACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCA .((.--------------..((((((((((......)).)).-))))))))(((((((((((...(((....))))))))).))))).(((((((((((......)))))..)))))).. ( -35.80) >DroSim_CAF1 12302 106 - 1 UGCC--------------CGGCGGUUCGCCUACUUUGGUUGAAAACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCA ....--------------.((((...)))).....(((((.....(((((((((((((((((...(((....))))))))).))))).))))))(((((......))))))))))..... ( -35.60) >DroEre_CAF1 17062 99 - 1 ---------------------UGGUUCGCCUACUUUGGUUGAAAACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCA ---------------------.((....)).....(((((.....(((((((((((((((((...(((....))))))))).))))).))))))(((((......))))))))))..... ( -31.80) >DroYak_CAF1 10816 120 - 1 UGGCAGGUGGCAGGUGGACGGUGGUUCGCCUACUUUGGUUGGAAACCGCGCGCCAAGCGAAAAAUUUGUUUGGAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCA .(((....((.((((((((....)))))))).)).((((..((..(((((((((((((..(((((((.....))))))).))))))).))))))..((....))...))..)))).))). ( -48.60) >consensus UGCC______________CGGCGGUUCGCCUACUUUGGUUGAAAACCGCGCGCCAACCGAAAAAUUUGUUUGCAGUUUUGGCUUGGCAGCGUGGAAAUGGAAAUGCAUUUGACCACGUCA ....................((((((.(((......)))....))))))..(((((((((((...(((....))))))))).))))).(((((((((((......)))))..)))))).. (-30.82 = -31.18 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:56 2006