
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,085,624 – 4,085,717 |
| Length | 93 |
| Max. P | 0.709458 |

| Location | 4,085,624 – 4,085,717 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.24 |
| Mean single sequence MFE | -17.50 |
| Consensus MFE | -12.34 |
| Energy contribution | -12.65 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4085624 93 + 22224390 UGCAAACACUCACA--------CACACUAGUGUGCACUCUUGUUUCACACAGAUACUGGCACACACACGCAUGCACAGAAAGGUGAGUGCUUACAAACCAU ((.((.(((((((.--------....((.(((((.((....))..)))))))...((((((..(....)..))).)))....))))))).)).))...... ( -22.60) >DroSim_CAF1 98873 97 + 1 UGCAAACACGCACACAC--GCACACACUAG--UGCACUCUUGUUUCACACAGAUACUGGCACACACACACAUGCACAGAAAGGUGAGUGCUUACAAACCAU .........((((.(((--((((......)--)))....................((((((..........))).)))....))).))))........... ( -20.40) >DroEre_CAF1 103564 97 + 1 UGCAAACACACAAACACACACACACACUAG--UACGCUCUUGUUUCACACAGAUACUGGCACAC--ACUCAUGCACAGAAAGGUGAGUGCUUACAAACCAU .............................(--(((((((.(.((((...(((...)))(((...--.....)))...)))).).)))))..)))....... ( -13.30) >DroYak_CAF1 96976 90 + 1 UGCAA-------AACACACACACACACUAG--UACACUCUUGUUUCACACAGAUACUGGCACAC--ACUCAUGCACAGAAAGGUGAGUGCUUACAAACCAU .....-------.................(--(((((((.(.((((...(((...)))(((...--.....)))...)))).).)))))..)))....... ( -13.70) >consensus UGCAAACAC_CAAACAC__ACACACACUAG__UACACUCUUGUUUCACACAGAUACUGGCACAC__ACUCAUGCACAGAAAGGUGAGUGCUUACAAACCAU .................................((((((.(.((((...(((...)))(((..........)))...)))).).))))))........... (-12.34 = -12.65 + 0.31)

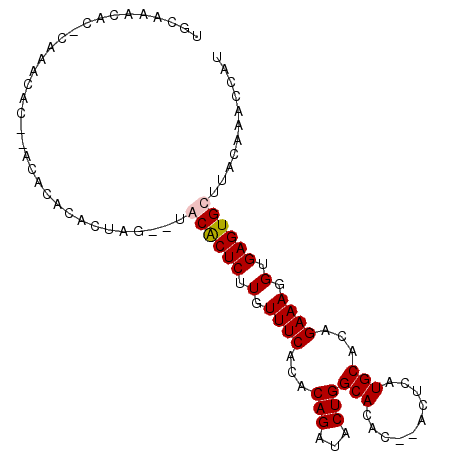
| Location | 4,085,624 – 4,085,717 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 86.24 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -19.45 |
| Energy contribution | -20.45 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4085624 93 - 22224390 AUGGUUUGUAAGCACUCACCUUUCUGUGCAUGCGUGUGUGUGCCAGUAUCUGUGUGAAACAAGAGUGCACACUAGUGUG--------UGUGAGUGUUUGCA ......(((((((((((((........((((((..(((((..((.((.((.....)).))..).)..)))))..)))))--------)))))))))))))) ( -34.00) >DroSim_CAF1 98873 97 - 1 AUGGUUUGUAAGCACUCACCUUUCUGUGCAUGUGUGUGUGUGCCAGUAUCUGUGUGAAACAAGAGUGCA--CUAGUGUGUGC--GUGUGUGCGUGUUUGCA ......((((((((((((((...(((.(((..(....)..)))))).....).)))).(((...(..((--(....)))..)--...)))..))))))))) ( -31.50) >DroEre_CAF1 103564 97 - 1 AUGGUUUGUAAGCACUCACCUUUCUGUGCAUGAGU--GUGUGCCAGUAUCUGUGUGAAACAAGAGCGUA--CUAGUGUGUGUGUGUGUUUGUGUGUUUGCA ......((((((((((((((...(((.((((....--..))))))).....).)))).(((((.(((((--(........)))))).)))))))))))))) ( -27.60) >DroYak_CAF1 96976 90 - 1 AUGGUUUGUAAGCACUCACCUUUCUGUGCAUGAGU--GUGUGCCAGUAUCUGUGUGAAACAAGAGUGUA--CUAGUGUGUGUGUGUGUU-------UUGCA ......(((((((((.((((...((((((((...(--((..(((((...))).))...)))...)))))--).))...).))).))).)-------))))) ( -23.90) >consensus AUGGUUUGUAAGCACUCACCUUUCUGUGCAUGAGU__GUGUGCCAGUAUCUGUGUGAAACAAGAGUGCA__CUAGUGUGUGU__GUGUGUG_GUGUUUGCA ......((((((((((((((...(((.((((........))))))).....).)))).(((..((......))..)))..............))))))))) (-19.45 = -20.45 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:26 2006