
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,083,683 – 4,083,779 |
| Length | 96 |
| Max. P | 0.960281 |

| Location | 4,083,683 – 4,083,779 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -14.25 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4083683 96 + 22224390 ------UGUGUCCCGCACUCGGAACAUCCGCUUACAUCCCCUCGUACACAUCC------------GCACACAUCGGAUGUUGCUUUGUUUGGGCAACUAAAUGCGCGUUUCCGA ------.(((.....)))((((((.((.(((............(((.((((((------------(.......))))))))))...((((((....))))))))).)))))))) ( -29.10) >DroSec_CAF1 96216 94 + 1 ------UGUGUUCCGCACUCGGAACAACCGCUAACAUC--CUCGUACACAUCC------------ACACACAUCGGAUGUUGCUUUGUUUGGGCAACUAAAUGCGCGUUUCCUA ------..(((((((....)))))))(((((.......--...(((.((((((------------.........)))))))))...((((((....))))))))).))...... ( -27.00) >DroSim_CAF1 97041 94 + 1 ------UGUGUUCCGCACUCGGAACAUCCGCUAACAUC--CUCGUACACAUCC------------GCACACAUCGGAUGUUGCUUUGUUUGGGCAACUAAAUGCGCGUUUCCGA ------.((((((((....)))))))).(((.......--...(((.((((((------------(.......))))))))))...((((((....))))))..)))....... ( -29.20) >DroEre_CAF1 101676 104 + 1 ------U-UGCCCCGCACUCGGAGCAUCCGCUUACAUC--CUUGUACACAUCCGCACACACAUCCGCACACAUCGGAUGUUGCUUUGUUUGGCCAACUAAAUG-GCGUUUCCGA ------.-..........((((((((..((..((((..--..))))..)....(((...(((((((.......))))))))))...)..))((((......))-))..)))))) ( -25.00) >DroYak_CAF1 95090 104 + 1 ------U-UGCCCCGCACUCGGAGCAUCCGCUUACAUC--CUUGUACACAUCCGCACUCACAUCUGCACACAUCGGAUGUUGCUUUGUUUGGCCAACUAAAUG-GCGUUUCCGA ------.-..........((((((((((((..((((..--..)))).......(((........)))......)))))))...........((((......))-))...))))) ( -24.40) >DroAna_CAF1 115182 106 + 1 GUUUGCUGUUUUCCGCA----AAAUAUCCGCAAACACC--CAC--ACACACCCACACACACACAUCCGCAUGUCGGAUGUUGCUUUGUUUGUGGAUCUAAAUUGGCAUUUCCGU ((((((((((((....)----)))))...))))))..(--((.--......(((((.(((((((((((.....)))))).))...))).)))))........)))......... ( -24.36) >consensus ______UGUGUCCCGCACUCGGAACAUCCGCUAACAUC__CUCGUACACAUCC____________GCACACAUCGGAUGUUGCUUUGUUUGGGCAACUAAAUGCGCGUUUCCGA ........(((.(((....))).)))..............................................(((((...(((...((((((....))))))..)))..))))) (-14.25 = -14.83 + 0.59)

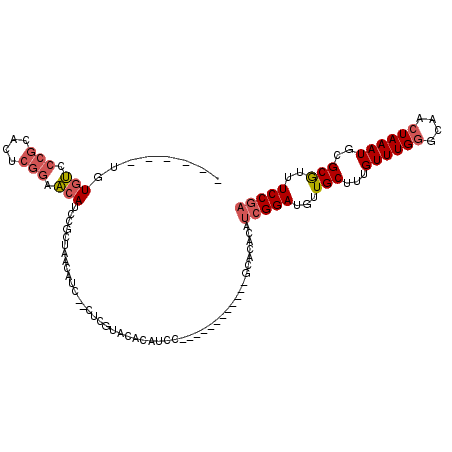
| Location | 4,083,683 – 4,083,779 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.64 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -18.21 |
| Energy contribution | -17.80 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4083683 96 - 22224390 UCGGAAACGCGCAUUUAGUUGCCCAAACAAAGCAACAUCCGAUGUGUGC------------GGAUGUGUACGAGGGGAUGUAAGCGGAUGUUCCGAGUGCGGGACACA------ ((((((.(((((.(((.(((.....)))))))).((((((....(((((------------(....))))))...))))))..)))....))))))(((.....))).------ ( -29.90) >DroSec_CAF1 96216 94 - 1 UAGGAAACGCGCAUUUAGUUGCCCAAACAAAGCAACAUCCGAUGUGUGU------------GGAUGUGUACGAG--GAUGUUAGCGGUUGUUCCGAGUGCGGAACACA------ ..(....)(.(((......))).).(((...(((((((((..((((..(------------....)..)))).)--)))))).)).)))((((((....))))))...------ ( -29.80) >DroSim_CAF1 97041 94 - 1 UCGGAAACGCGCAUUUAGUUGCCCAAACAAAGCAACAUCCGAUGUGUGC------------GGAUGUGUACGAG--GAUGUUAGCGGAUGUUCCGAGUGCGGAACACA------ .((....)).(((......))).........(((((((((..((((..(------------....)..)))).)--)))))).))...(((((((....)))))))..------ ( -31.70) >DroEre_CAF1 101676 104 - 1 UCGGAAACGC-CAUUUAGUUGGCCAAACAAAGCAACAUCCGAUGUGUGCGGAUGUGUGUGCGGAUGUGUACAAG--GAUGUAAGCGGAUGCUCCGAGUGCGGGGCA-A------ ..(....)((-((......))))........((.((((((....((((((.((.((....)).)).)))))).)--)))))..))...(((((((....)))))))-.------ ( -39.00) >DroYak_CAF1 95090 104 - 1 UCGGAAACGC-CAUUUAGUUGGCCAAACAAAGCAACAUCCGAUGUGUGCAGAUGUGAGUGCGGAUGUGUACAAG--GAUGUAAGCGGAUGCUCCGAGUGCGGGGCA-A------ ..(....)((-((......))))........((.((((((..((((..((..(((....)))..))..)))).)--)))))..))...(((((((....)))))))-.------ ( -39.50) >DroAna_CAF1 115182 106 - 1 ACGGAAAUGCCAAUUUAGAUCCACAAACAAAGCAACAUCCGACAUGCGGAUGUGUGUGUGUGGGUGUGU--GUG--GGUGUUUGCGGAUAUUU----UGCGGAAAACAGCAAAC .((.(((((((....((.(((((((.(((.....(((((((.....))))))).))).))))))).)).--...--))))))).)).....((----(((........))))). ( -39.40) >consensus UCGGAAACGCGCAUUUAGUUGCCCAAACAAAGCAACAUCCGAUGUGUGC____________GGAUGUGUACGAG__GAUGUAAGCGGAUGUUCCGAGUGCGGAACACA______ .......(((.((((..(((.....)))...(((.(((((.....................)))))))).......))))...)))..(((((((....)))))))........ (-18.21 = -17.80 + -0.41)
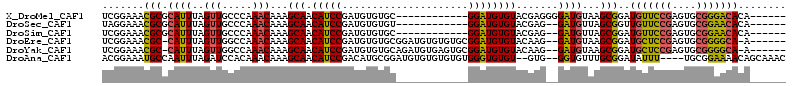


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:24 2006