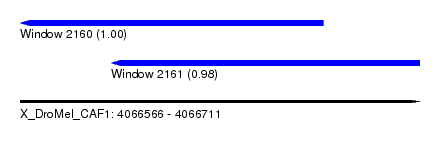
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,066,566 – 4,066,711 |
| Length | 145 |
| Max. P | 0.999547 |

| Location | 4,066,566 – 4,066,676 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.23 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.16 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4066566 110 - 22224390 GCCAAUGCAUCAACGUUGGCAUCAACGACCUUGGUAACUUGAUUGUUAUUUCCAACAAUAUCAACGCUAGGAUCGACUAUUAAAAAAAU-------GAUUUCAAGUACAUAAGUAAC (((((((......))))))).....(((.((((((......((((((......))))))......)))))).)))..............-------..................... ( -22.60) >DroSec_CAF1 79415 110 - 1 GCCAAUGCAUCAACGUUGGCAUCAACGACCUUGGUAACUUGGUUGUUAUUUCCAACAAUAUCAACACUGGGGUCGACUAUAACAAAAGU-------AAUUUCAAGUUCAUAGGUGAC (((((((......))))))).(((.((((((..((......((((((......))))))......))..)))))).(((((((.(((..-------..)))...))).)))).))). ( -32.00) >DroSim_CAF1 78403 110 - 1 GCCAAUGCAUCAACGUUGGCAUCAACGACCUUGGUAACUUGGUUGUUAUUUCCAACAAUAUCAACACUGGGGUCGACUAUAACAAAAGU-------AAUUUCAAGUUCAUAGGUGAC (((((((......))))))).(((.((((((..((......((((((......))))))......))..)))))).(((((((.(((..-------..)))...))).)))).))). ( -32.00) >DroEre_CAF1 78784 110 - 1 GCCAAUGCAUCAACGUUGGCAUCAGCGACCUUGGUAAUUUGGUUGUUAUUUCCAACAAUAUCAACGCUGGGGUCGACUAUUAAAUAAGU-------UAUUUAAGGUAUAUAAGUAAC (((((((......)))))))...((((((((..((......((((((......))))))......))..)))))).)).........((-------((((((.......)))))))) ( -33.10) >DroYak_CAF1 78482 117 - 1 GCCAACACAUCAACGUUGGCAUCAACGACCUUGGAAAUUUGGUUGUUAUUUCCAACAAUAUCAACGCUGGGGUCGACUAUUUUAUAAUUGUAUAAAUAUUUGAAGUACAUAAGUAAA ((((((........)))))).....((((((..(.......((((((......)))))).......)..)))))).....(((((...(((((...........)))))...))))) ( -30.44) >consensus GCCAAUGCAUCAACGUUGGCAUCAACGACCUUGGUAACUUGGUUGUUAUUUCCAACAAUAUCAACGCUGGGGUCGACUAUUAAAAAAGU_______AAUUUCAAGUACAUAAGUAAC (((((((......))))))).....((((((((((......((((((......))))))......)))))))))).......................................... (-26.20 = -26.16 + -0.04)


| Location | 4,066,599 – 4,066,711 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.27 |
| Mean single sequence MFE | -34.94 |
| Consensus MFE | -24.08 |
| Energy contribution | -23.57 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4066599 112 - 22224390 -----CAGUUCCAGUAAUUUUGCUAAGGACCGGGACAUUAGCCAAUGCAUCAACGUUGGCAUCAACGACCUUGGUAACUUGAUUGUUAUUUCCAAC---AAUAUCAACGCUAGGAUCGAC -----..(((((.((..((.....))..)).)))))....(((((((......))))))).....(((.((((((......((((((......)))---)))......)))))).))).. ( -30.40) >DroSec_CAF1 79448 112 - 1 -----UAGCUCCAGUUAUUUGGCUAAGGACCGGGACAUUAGCCAAUGCAUCAACGUUGGCAUCAACGACCUUGGUAACUUGGUUGUUAUUUCCAAC---AAUAUCAACACUGGGGUCGAC -----..((((((((...(((((((((........).)))))))).........(((((.(..(((((((..(....)..)))))))..).)))))---.........)))))))).... ( -36.60) >DroSim_CAF1 78436 112 - 1 -----UAGCUCCAGUUAUUUGGCUAAGGACCGGGACAUUAGCCAAUGCAUCAACGUUGGCAUCAACGACCUUGGUAACUUGGUUGUUAUUUCCAAC---AAUAUCAACACUGGGGUCGAC -----..((((((((...(((((((((........).)))))))).........(((((.(..(((((((..(....)..)))))))..).)))))---.........)))))))).... ( -36.60) >DroEre_CAF1 78817 112 - 1 -----CAGCUCCAGUUAUUUUGCUAAGGACCGGGAUAUUAGCCAAUGCAUCAACGUUGGCAUCAGCGACCUUGGUAAUUUGGUUGUUAUUUCCAAC---AAUAUCAACGCUGGGGUCGAC -----(.((((((((((..(((((((((..((.(((....(((((((......))))))))))..)).)))))))))..))((((.((((......---)))).)))))))))))).).. ( -37.00) >DroYak_CAF1 78522 112 - 1 -----CAGCUCCAGUUAUUUGGCUAAGGACCGGGGCAUUUGCCAACACAUCAACGUUGGCAUCAACGACCUUGGAAAUUUGGUUGUUAUUUCCAAC---AAUAUCAACGCUGGGGUCGAC -----..(((((.(((.((.....)).))).)))))...(((((((........)))))))....((((((..(.......((((((......)))---))).......)..)))))).. ( -38.44) >DroMoj_CAF1 116261 112 - 1 UGUAACCGUUCCAGCCCUGUUGGUCAGCACAUUGGCAUCCACAGCCGCAGGCACAUCGAGACCCACAAGUUGGCCAAAGAAAUUGUUAUUUCCACUAACGAUAUU---UUUGGGA----- ..........(((((..(((.((((.((......)).......(((...))).......)))).))).)))))(((((((.(((((((.......))))))).))---)))))..----- ( -30.60) >consensus _____CAGCUCCAGUUAUUUGGCUAAGGACCGGGACAUUAGCCAAUGCAUCAACGUUGGCAUCAACGACCUUGGUAACUUGGUUGUUAUUUCCAAC___AAUAUCAACGCUGGGGUCGAC .......((((((((......(((((((.....((.....(((((((......))))))).)).....)))))))......((((.((((.........)))).)))))))))))).... (-24.08 = -23.57 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:07:16 2006