
| Sequence ID | X_DroMel_CAF1 |
|---|---|
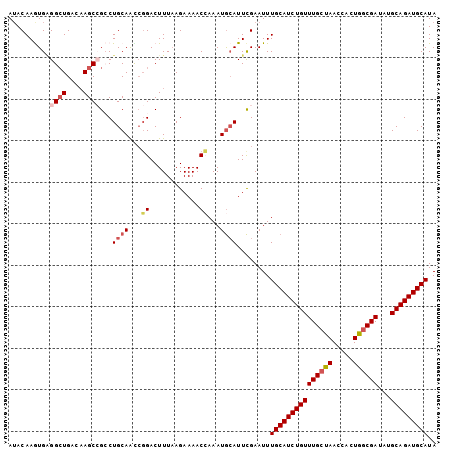
| Location | 4,022,982 – 4,023,119 |
| Length | 137 |
| Max. P | 0.703427 |

| Location | 4,022,982 – 4,023,085 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 90.87 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -19.52 |
| Energy contribution | -20.82 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
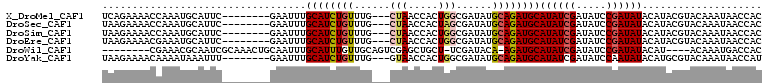
>X_DroMel_CAF1 4022982 103 + 22224390 AUACAAGUGAGGCUGAAAAGCCGCCUGCAAUCGGACUUUCAGAAAACCAAAUGCAUUCGAAUUUGCAUCUGUUUGCUAACCACUGGCGAUAUGCAGAUGCAUA ..........((((....))))(..((((...((............))...))))..).....(((((((((((((((.....))))))...))))))))).. ( -29.00) >DroSec_CAF1 36886 103 + 1 AUACAAGUGAGGCUGACAAGCCGCCUGCAACCGGACUUUAAGAAAACCAAAUGCAUUCGAAUUUGCAUCUGUUUGCUAACCACUAGCGAUAUGCAGAUGCAUA ..........((((....))))(..((((...((............))...))))..).....(((((((((((((((.....))))))...))))))))).. ( -27.80) >DroSim_CAF1 35837 103 + 1 AUACAAGUGAGGCUGACAAGCCGCCUGCAACCGGACUUUAAGAAAACCAAAUGCAUUCGAAUUUGCAUCUGUUUGCUAACCACUGGCGAUAUGCAGAUGCAUA ..........((((....))))(..((((...((............))...))))..).....(((((((((((((((.....))))))...))))))))).. ( -27.50) >DroEre_CAF1 36512 103 + 1 AAACAAAUGGGGAUGACCAGCAGACUGCAGCACGACUUUAAGAAAACGAAAUGCAUUCGAAUUUGCAUCUGUUUGCUAACCACUGGCGAUAUGCAGAUGCAUA .......(((......)))...((.((((...((............))...)))).)).....(((((((((((((((.....))))))...))))))))).. ( -26.40) >DroYak_CAF1 34706 103 + 1 AUACAAAUGUGGCUGGCAAGCAGACUGCAAUCGGACUUUAAGAAAACAAAAUAAAUUUGAAUUUGCAUCUGUUUGGUAACCACUGGCGAUAUGCAGAUGCAUA ........((((.((.((((((((.((((((((((.((((...........)))))))))..))))))))))))).)).))))......(((((....))))) ( -28.00) >consensus AUACAAGUGAGGCUGACAAGCCGCCUGCAACCGGACUUUAAGAAAACCAAAUGCAUUCGAAUUUGCAUCUGUUUGCUAACCACUGGCGAUAUGCAGAUGCAUA ..........((((....))))...((((...((............))...))))........(((((((((((((((.....))))))...))))))))).. (-19.52 = -20.82 + 1.30)


| Location | 4,023,020 – 4,023,119 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -13.84 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
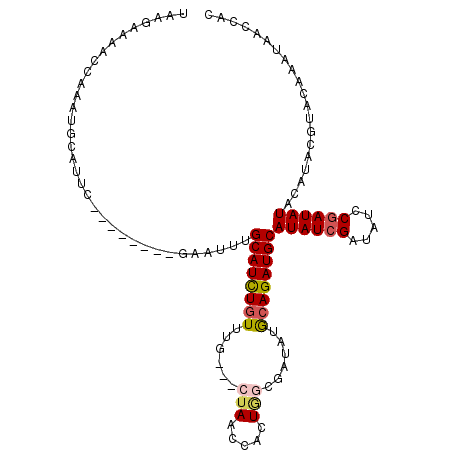
>X_DroMel_CAF1 4023020 99 + 22224390 UCAGAAAACCAAAUGCAUUC--------GAAUUUGCAUCUGUUUG---CUAACCACUGGCGAUAUGCAGAUGCAUAUCGAUAUCCGAUAUACAUACGUACAAAUAACCAC .............(((....--------......(((((((((((---(((.....))))))...))))))))((((((.....))))))......)))........... ( -23.20) >DroSec_CAF1 36924 99 + 1 UAAGAAAACCAAAUGCAUUC--------GAAUUUGCAUCUGUUUG---CUAACCACUAGCGAUAUGCAGAUGCAUAUCGAUAUCCGAUAUACAUACGUACAAAUAACCAC .............(((....--------......(((((((((((---(((.....))))))...))))))))((((((.....))))))......)))........... ( -23.50) >DroSim_CAF1 35875 99 + 1 UAAGAAAACCAAAUGCAUUC--------GAAUUUGCAUCUGUUUG---CUAACCACUGGCGAUAUGCAGAUGCAUAUCGAUAUCCGAUAUACAUACGUACAAAUAACCAC .............(((....--------......(((((((((((---(((.....))))))...))))))))((((((.....))))))......)))........... ( -23.20) >DroEre_CAF1 36550 99 + 1 UAAGAAAACGAAAUGCAUUC--------GAAUUUGCAUCUGUUUG---CUAACCACUGGCGAUAUGCAGAUGCAUAUCGAUAUCCGAUAUACAUACGUACAAAUAACCAC .......(((..........--------......(((((((((((---(((.....))))))...))))))))((((((.....)))))).....)))............ ( -24.10) >DroWil_CAF1 44983 96 + 1 --------CGAAACGCAAUCGCAAACUGCAAUUUGCAUUUGUUGCAGUCGAGCUGCU-UCGAUACA-AGAUGCAUAUCGAUAUCCGAUAUACAU----ACAAAUGACCAC --------((((..(((.(((...((((((((........)))))))))))..))))-))).....-..(((.((((((.....)))))).)))----............ ( -24.30) >DroYak_CAF1 34744 99 + 1 UAAGAAAACAAAAUAAAUUU--------GAAUUUGCAUCUGUUUG---GUAACCACUGGCGAUAUGCAGAUGCAUAUCGAUAUCCAAUAUACAUGCGUACAAAUAACCAU ................((((--------(....(((((.((((((---(..........((((((((....))))))))....)))).))).)))))..)))))...... ( -20.54) >consensus UAAGAAAACCAAAUGCAUUC________GAAUUUGCAUCUGUUUG___CUAACCACUGGCGAUAUGCAGAUGCAUAUCGAUAUCCGAUAUACAUACGUACAAAUAACCAC ..................................((((((((......(((.....)))......))))))))((((((.....)))))).................... (-13.84 = -14.28 + 0.45)
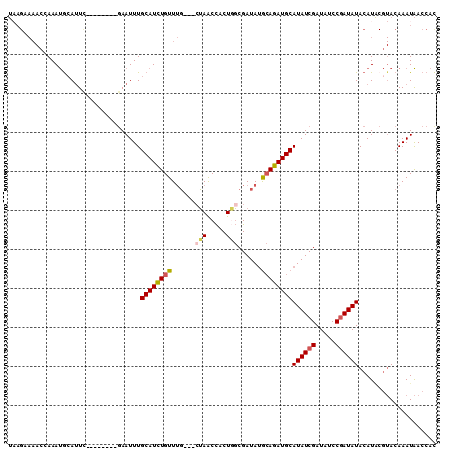
| Location | 4,023,020 – 4,023,119 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.77 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4023020 99 - 22224390 GUGGUUAUUUGUACGUAUGUAUAUCGGAUAUCGAUAUGCAUCUGCAUAUCGCCAGUGGUUAG---CAAACAGAUGCAAAUUC--------GAAUGCAUUUGGUUUUCUGA .((((.((.((((.(.((((((((((.....))))))))))))))).)).))))....((((---.(((((((((((.....--------...))))))).)))).)))) ( -30.00) >DroSec_CAF1 36924 99 - 1 GUGGUUAUUUGUACGUAUGUAUAUCGGAUAUCGAUAUGCAUCUGCAUAUCGCUAGUGGUUAG---CAAACAGAUGCAAAUUC--------GAAUGCAUUUGGUUUUCUUA (((((....((((.(.((((((((((.....))))))))))))))).)))))..........---....((((((((.....--------...))))))))......... ( -27.00) >DroSim_CAF1 35875 99 - 1 GUGGUUAUUUGUACGUAUGUAUAUCGGAUAUCGAUAUGCAUCUGCAUAUCGCCAGUGGUUAG---CAAACAGAUGCAAAUUC--------GAAUGCAUUUGGUUUUCUUA .((((.((.((((.(.((((((((((.....))))))))))))))).)).))))........---....((((((((.....--------...))))))))......... ( -28.60) >DroEre_CAF1 36550 99 - 1 GUGGUUAUUUGUACGUAUGUAUAUCGGAUAUCGAUAUGCAUCUGCAUAUCGCCAGUGGUUAG---CAAACAGAUGCAAAUUC--------GAAUGCAUUUCGUUUUCUUA .((((.((.((((.(.((((((((((.....))))))))))))))).)).))))......((---.(((((((((((.....--------...))))))).)))).)).. ( -27.80) >DroWil_CAF1 44983 96 - 1 GUGGUCAUUUGU----AUGUAUAUCGGAUAUCGAUAUGCAUCU-UGUAUCGA-AGCAGCUCGACUGCAACAAAUGCAAAUUGCAGUUUGCGAUUGCGUUUCG-------- (..(((((((((----((((((((((.....))))))))))..-((((((((-......)))).))))))))))((((((....)))))))))..)......-------- ( -31.90) >DroYak_CAF1 34744 99 - 1 AUGGUUAUUUGUACGCAUGUAUAUUGGAUAUCGAUAUGCAUCUGCAUAUCGCCAGUGGUUAC---CAAACAGAUGCAAAUUC--------AAAUUUAUUUUGUUUUCUUA .(((((((.((....)).)))((((((....((((((((....))))))))))))))...))---))...(((.(((((...--------........)))))..))).. ( -21.40) >consensus GUGGUUAUUUGUACGUAUGUAUAUCGGAUAUCGAUAUGCAUCUGCAUAUCGCCAGUGGUUAG___CAAACAGAUGCAAAUUC________GAAUGCAUUUGGUUUUCUUA .((((.((.((((.(.((((((((((.....))))))))))))))).)).))))............(((((((((((................))))))).))))..... (-19.21 = -19.77 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:58 2006