
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 4,016,219 – 4,016,348 |
| Length | 129 |
| Max. P | 0.950420 |

| Location | 4,016,219 – 4,016,321 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -25.94 |
| Energy contribution | -27.82 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4016219 102 + 22224390 -CUAGCCACCGCCUGUUCGUUCCACUGGACUCUCCCGGGGGAGUGGAUUUCGUCUCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCC -...(((.(((...(((((((((.((((......)))).)))))))))........((..((((((.....))))))..))))))))................ ( -31.20) >DroSec_CAF1 30469 99 + 1 AUUUGCCACCGCCUGU----UCCCCUGGACACUCCCGGGGGAGUGGAUUUCGUCUCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCC ....(((.(((.(..(----((((((((......)))))))))..)..........((..((((((.....))))))..))))))))................ ( -38.40) >DroSim_CAF1 29331 99 + 1 GUUUGCCACCGCCUGU----UCCCCUGGACACUCCCGGGGGAGUGGAUUUCGUCUCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCC ....(((.(((.(..(----((((((((......)))))))))..)..........((..((((((.....))))))..))))))))................ ( -38.40) >DroEre_CAF1 28928 86 + 1 GUUUUCCACCGCCUUU-----------------CCCGGGGGAGUGGAUUUCGUCUCUGUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCC ....((((((.(((..-----------------...))).).)))))..(((.((((((.((((((.....))))))...))))))....))).......... ( -28.70) >DroYak_CAF1 28713 98 + 1 GUUUUCCACCGCCUUU----UUCCCU-GACUUACCCGGGGGAGUGGAUUUCGUCCCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUUC (...(((((.......----((((((-(.......))))))))))))...)(((((((..((((((.....))))))..)).)))))................ ( -32.81) >consensus GUUUGCCACCGCCUGU____UCCCCUGGACACUCCCGGGGGAGUGGAUUUCGUCUCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCC ....................((((((((......))))))))((((.(..((((......((((((.....))))))..))))..)))))............. (-25.94 = -27.82 + 1.88)

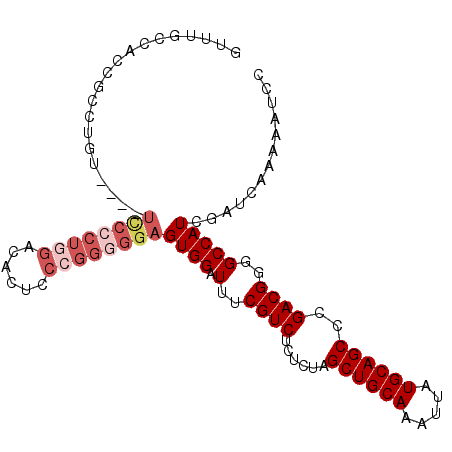
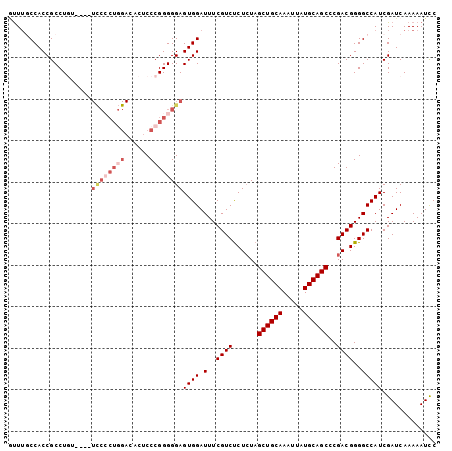
| Location | 4,016,219 – 4,016,321 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.08 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4016219 102 - 22224390 GGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGAGACGAAAUCCACUCCCCCGGGAGAGUCCAGUGGAACGAACAGGCGGUGGCUAG- .((((...))))..((((((((((.(((((((.....)))))))............((((((((....)))(....).))))).......))))).))))).- ( -37.10) >DroSec_CAF1 30469 99 - 1 GGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGAGACGAAAUCCACUCCCCCGGGAGUGUCCAGGGGA----ACAGGCGGUGGCAAAU .((((...))))....((((((((.(((((((.....))))))).................(((((.(((....))).)))))----...))))).))).... ( -38.30) >DroSim_CAF1 29331 99 - 1 GGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGAGACGAAAUCCACUCCCCCGGGAGUGUCCAGGGGA----ACAGGCGGUGGCAAAC .((((...))))....((((((((.(((((((.....))))))).................(((((.(((....))).)))))----...))))).))).... ( -38.30) >DroEre_CAF1 28928 86 - 1 GGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUACAGAGACGAAAUCCACUCCCCCGGG-----------------AAAGGCGGUGGAAAAC (((((((...((((((....)))).(((((((.....)))))))...))...)))))))..(((.(((..-----------------.....))).))).... ( -31.60) >DroYak_CAF1 28713 98 - 1 GAAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGGGACGAAAUCCACUCCCCCGGGUAAGUC-AGGGAA----AAAGGCGGUGGAAAAC ...(((((.((((....((((.((.(((((((.....))))))).)).)(((...((((........))))..)))-.)))..----.....)))).))))). ( -32.20) >consensus GGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGAGACGAAAUCCACUCCCCCGGGAGAGUCCAGGGGA____ACAGGCGGUGGCAAAC (((((((...((((((....)))).(((((((.....)))))))...))...))))))).(..((((.......((.....)).......)).))..)..... (-27.74 = -28.08 + 0.34)



| Location | 4,016,258 – 4,016,348 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.08 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4016258 90 + 22224390 GAGUGGAUUUCGUCUCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCCGUAUGCAGGGAGCAGCUGGGUCGGAUU ((.(((.(..((((......((((((.....))))))..))))..)))))).......((((((.((.(((.......))).)))))))) ( -30.10) >DroSec_CAF1 30505 89 + 1 GAGUGGAUUUCGUCUCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCCGUAUGCAG-GAGCAGCUGGGUCGGAUU ((.(((.(..((((......((((((.....))))))..))))..)))))).......((((((.((.(((-......))).)))))))) ( -30.60) >DroSim_CAF1 29367 89 + 1 GAGUGGAUUUCGUCUCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCCGUAUGCAG-GAGCAGCUGGGUCGGAUU ((.(((.(..((((......((((((.....))))))..))))..)))))).......((((((.((.(((-......))).)))))))) ( -30.60) >DroEre_CAF1 28951 89 + 1 GAGUGGAUUUCGUCUCUGUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCCGUACGCAG-GAGCAGCUGGGACGGAUU ((.(((.(..((((......((((((.....))))))..))))..)))))).......(((((((.(.(((-......))).)))))))) ( -32.10) >DroYak_CAF1 28748 89 + 1 GAGUGGAUUUCGUCCCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUUCGUACGCAG-GAGCAGCUGGUUCGGAUU (((.....)))((((..((((((((......(((.((((....))))...(((........)))...))).-..))))))))...)))). ( -29.20) >consensus GAGUGGAUUUCGUCUCUCUAGCUGCAAAUUAUGCAGCCCGACGGGGCCAUCGAUCAAAAAUCCGUAUGCAG_GAGCAGCUGGGUCGGAUU ((.(((.(..((((......((((((.....))))))..))))..)))))).......((((((.((.(((.......))).)))))))) (-26.96 = -27.08 + 0.12)



| Location | 4,016,258 – 4,016,348 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 96.64 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 4016258 90 - 22224390 AAUCCGACCCAGCUGCUCCCUGCAUACGGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGAGACGAAAUCCACUC .............(((.....)))...(((((((...((((((....)))).(((((((.....)))))))...))...))))))).... ( -27.20) >DroSec_CAF1 30505 89 - 1 AAUCCGACCCAGCUGCUC-CUGCAUACGGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGAGACGAAAUCCACUC .............(((..-..)))...(((((((...((((((....)))).(((((((.....)))))))...))...))))))).... ( -27.20) >DroSim_CAF1 29367 89 - 1 AAUCCGACCCAGCUGCUC-CUGCAUACGGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGAGACGAAAUCCACUC .............(((..-..)))...(((((((...((((((....)))).(((((((.....)))))))...))...))))))).... ( -27.20) >DroEre_CAF1 28951 89 - 1 AAUCCGUCCCAGCUGCUC-CUGCGUACGGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUACAGAGACGAAAUCCACUC ..(((((.((((......-))).).)))))((((((..(((((....)))))(((((((.....))))))).))))))............ ( -29.40) >DroYak_CAF1 28748 89 - 1 AAUCCGAACCAGCUGCUC-CUGCGUACGAAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGGGACGAAAUCCACUC ....((.(((((......-))).)).))..........(((((....)))))(((((((.....))))))).((((((.....))).))) ( -25.90) >consensus AAUCCGACCCAGCUGCUC_CUGCAUACGGAUUUUUGAUCGAUGGCCCCGUCGGGCUGCAUAAUUUGCAGCUAGAGAGACGAAAUCCACUC .............(((.....)))...(((((((...((((((....)))).(((((((.....)))))))...))...))))))).... (-25.80 = -25.60 + -0.20)

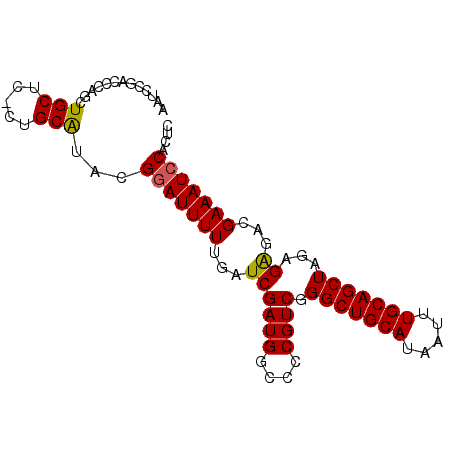
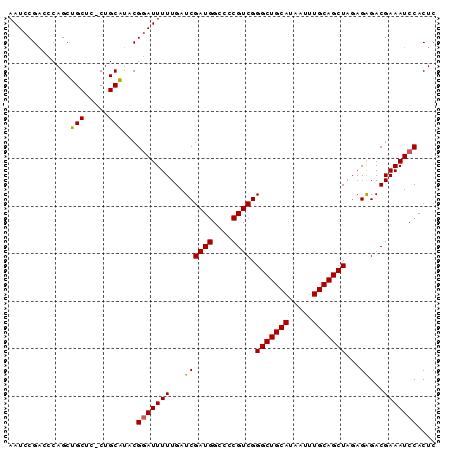
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:54 2006