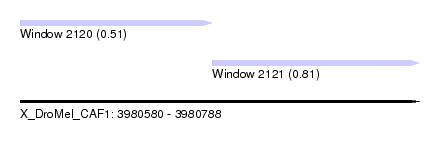
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,980,580 – 3,980,788 |
| Length | 208 |
| Max. P | 0.812623 |

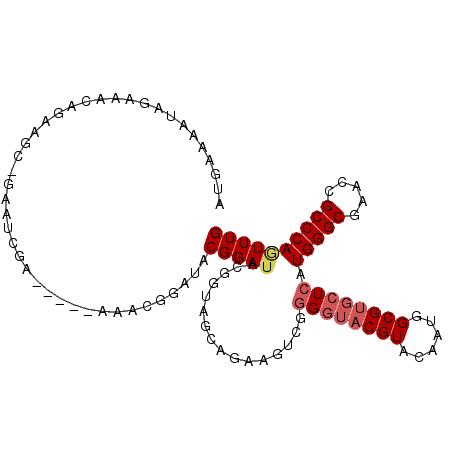
| Location | 3,980,580 – 3,980,680 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -18.09 |
| Energy contribution | -18.82 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3980580 100 + 22224390 AUGAAAAUAGAAACAGAAGC-GAAUCGA-----AAACGGAUACGGAUACGGUAGCGGAAGUCGGGGUACGUACAAUGGCGUGCUCAUGGGCGAACCGCCCAGUUUG ..........((((....((-..((((.-----(..((....))..).)))).))........((((((((......)))))))).((((((...)))))))))). ( -28.80) >DroSec_CAF1 2617 100 + 1 AUGAAAAUAGAAACAGAAGC-GAAUCGA-----AAACGGAUACGGAUACGGUAGCGGAAGUCGGGGUACGUACAAUGGCGUGCUCAUGGGCGAACCGCCCAGUUUG ..........((((....((-..((((.-----(..((....))..).)))).))........((((((((......)))))))).((((((...)))))))))). ( -28.80) >DroSim_CAF1 553 100 + 1 AUGAAAAUAGAAACAGAAGC-GAAUCGA-----AAACGGAUACGGAUACGGUAGCGGAAGUCGGGGUACGUACAAUGGCGUGCUCAUGGGCGAACCGCCCAGUUUG ..........((((....((-..((((.-----(..((....))..).)))).))........((((((((......)))))))).((((((...)))))))))). ( -28.80) >DroEre_CAF1 473 105 + 1 AUGAAAAUAGCAACGGAAAC-GAAUCGAAUCAAAAACGGAUACGGAUACGGAUACAGAAGUCGGGGUACGUACAAUGGCGUGCUCAUGGGCGAACCGCCCAGUUUG .............((....)-)..(((.(((.......))).)))...(((((..........((((((((......)))))))).((((((...))))))))))) ( -27.70) >DroYak_CAF1 564 103 + 1 AUGAAAAUAGAAUAAGAAACAGAAACGAAC---GAACGGAUACGGAUACGGAUAGAGAAGUCGGGGUACGUACAAUGGCGUGCUCAUGGGCGAACAGCCCAAUUUG ...................((((..(((..---.........((....))..........)))((((((((......)))))))).(((((.....))))).)))) ( -24.00) >DroAna_CAF1 787 89 + 1 ACGGAAACUGAAGCAG----------GA-----UAACUGAAGCGGAGAGAGUGGUAGAG--AGAGGUACGUACAAUGGCGAUGUCAUGGGCGAACCGCCCAGUUUG ....((((((..((..----------..-----...(((..((.......))..)))..--...(((.(((...(((((...)))))..))).))))).)))))). ( -21.50) >consensus AUGAAAAUAGAAACAGAAGC_GAAUCGA_____AAACGGAUACGGAUACGGUAGCAGAAGUCGGGGUACGUACAAUGGCGUGCUCAUGGGCGAACCGCCCAGUUUG ..........................................(((((................((((((((......)))))))).(((((.....)))))))))) (-18.09 = -18.82 + 0.72)


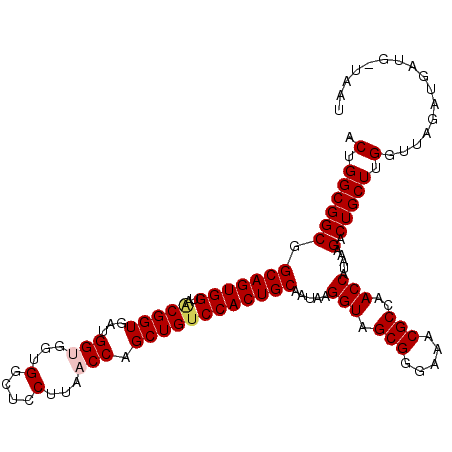
| Location | 3,980,680 – 3,980,788 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.02 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -32.92 |
| Energy contribution | -33.40 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3980680 108 + 22224390 ACGGGCGGCGGCAGUGGCAACGGUGACGGCGGUGGCUCCUUUACCAGCUGGCCACUGCAAUAAGGUAGCGGAAAACGCCAACCGUAAAGACUGCUUGGUUAGAUGGU-----U .((((((((.((((((((..((....))(((((((.....))))).))..)))))))).....(((.(((.....)))..))).....).)))))))..........-----. ( -42.90) >DroVir_CAF1 619 113 + 1 ACUGGCGGCGGCAGUGGUAACGGUGAUGGUGGUGGCUCCUUAACCAGCUGUCCACUGCAAUAAGGUAGCGGGAAACGCCAACCAUAAAGACUGCUUGGUUAGAUGAUGGUAAU ..((((.((.(((((((..(((((..((((...(....)...)))))))))))))))).........)).(....)))))(((((...((((....)))).....)))))... ( -36.50) >DroGri_CAF1 639 113 + 1 ACUGGCGGCGGCAGUGGUAACGGUGAUGGUGGUGGCUCCUUAACCAGCUGUCCACUGCAAUAAGGUAGCGGGAAACGCCAACCAUAAAGACUGCUUGGUUAGAUGAUGAUAAU .((((((((((((((((..(((((..((((...(....)...)))))))))))))))).....(((.(((.....)))..)))........))))..)))))........... ( -36.20) >DroWil_CAF1 8787 105 + 1 ACUGGCGGCGGCAGUGGUAGCGGUGACGGUGGUGGCUCCUUAACCAGCUGUCCACUGCAAUAAGGUAGCGGAAAACGCCAACCAUAAAGACUGCUUGGUUAAAU--------U .(.((((((.((((((((((((....)(((...(....)...))).)))).))))))).....(((.(((.....)))..))).....).))))).).......--------. ( -38.80) >DroMoj_CAF1 625 113 + 1 ACUGGCGGCGGCAGUGGCAACGGUGAUGGCGGCGGCUCCUUAACCAGCUGUCCACUGCAAUAAGGUAGCGGGAAACGCCAACCAUAAAGACUGCUUGGUUAGAUGUUGAUAAU .(((((.(.(((((((....)(((..(((((((((((........))))))((.((((......)))).))....))))))))......))))))).)))))........... ( -39.10) >consensus ACUGGCGGCGGCAGUGGUAACGGUGAUGGUGGUGGCUCCUUAACCAGCUGUCCACUGCAAUAAGGUAGCGGGAAACGCCAACCAUAAAGACUGCUUGGUUAGAUGAUG_UAAU .(.((((((.(((((((..(((((...(((...(....)...))).)))))))))))).....(((.(((.....)))..))).....).))))).)................ (-32.92 = -33.40 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:41 2006