
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,953,387 – 3,953,530 |
| Length | 143 |
| Max. P | 0.814187 |

| Location | 3,953,387 – 3,953,491 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.69 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3953387 104 - 22224390 AAUCUUUGUUCUGAUAAUGCUGCU--GAAUUGCUCACAAUUCUUUAUCGGUACCAAAAUAACAGCAAUGAGCCAAUAAUGAGAAAACGGCAACUGAACAAUAAAGA-------------- .....((((((.(....(((((..--((((((....))))))......(....).......)))))....(((..............)))..).))))))......-------------- ( -18.64) >DroSim_CAF1 3822 120 - 1 AAUCUUUGUUCUGAUAAUUCUGCUGUGAAUUGCUCACAAUUCUUUAUCGGGACCAAAAUAACAGCGGUGAGCCAAUAAUUAGAAAACGGCAACUGAACAAUAAAGAAUAUCUGCUCCAGA ..(((((((((((((((......(((((.....))))).....))))))))))...........((((..(((..............))).))))......)))))...((((...)))) ( -24.84) >DroEre_CAF1 3803 118 - 1 AAUCUUUGUCCUGAUAAUGCUGAU--GAAUUGCCCACAAUUCUUUAUCAGCACCAAAAUAACAGCGGUGAGCCAAUAAUAAGAAAACGGCAACUGAACAAUAAAGAAUAUCUGCUCUAGA ..(((((((..((....(((((((--((((((....))))))...)))))))............((((..(((..............))).))))..))))))))).............. ( -23.84) >DroYak_CAF1 3713 118 - 1 AGUCUUUGUUCUGAUAAUGCCAAU--GAAUUGGCCACAAUUCUUUAUCGGUACCAAAAUAACAGCGGUGAGCCAAUAAUGAGAAAACGGCAACUGAACAAUAAAGAAUAUCUGCUCUAGA ..((((((((.......((((...--((((((....)))))).((((.((((((...........)))..))).)))).........)))).......)))))))).............. ( -22.34) >consensus AAUCUUUGUUCUGAUAAUGCUGAU__GAAUUGCCCACAAUUCUUUAUCGGUACCAAAAUAACAGCGGUGAGCCAAUAAUGAGAAAACGGCAACUGAACAAUAAAGAAUAUCUGCUCUAGA ..((((((((.......(((((((..((((((....))))))...)))))))............((((..(((..............))).))))...)))))))).............. (-20.23 = -20.42 + 0.19)

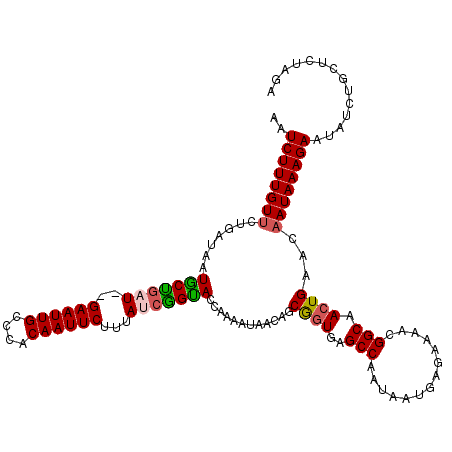
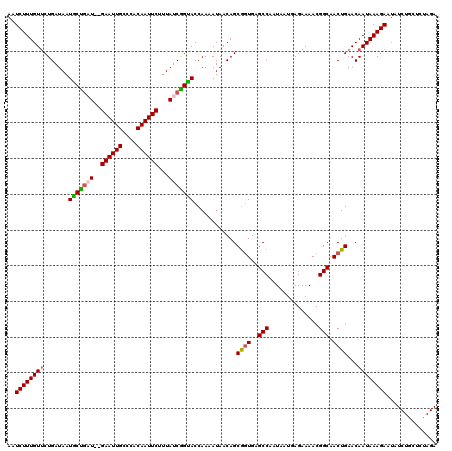
| Location | 3,953,413 – 3,953,530 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -18.19 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3953413 117 + 22224390 CAUUAUUGGCUCAUUGCUGUUAUUUUGGUACCGAUAAAGAAUUGUGAGCAAUUC--AGCAGCAUUAUCAGAACAAAGAUUGGCGGGCCAAAUAAUGUUGAAUUAAUCAUCA-UCACUUAC ((((((((((((..(((((((...(((....)))....((((((....))))))--)))))))..(((........)))....))))).)))))))...............-........ ( -28.40) >DroSim_CAF1 3862 119 + 1 AAUUAUUGGCUCACCGCUGUUAUUUUGGUCCCGAUAAAGAAUUGUGAGCAAUUCACAGCAGAAUUAUCAGAACAAAGAUUAGCGGGCCAAAUAAUGUUGAAUUAAUCAUCA-UCAGUUGC .....(((((...((((((...(((((.((..(((((....(((((((...))))))).....))))).)).)))))..)))))))))))...(((.(((.....))).))-)....... ( -26.90) >DroEre_CAF1 3843 107 + 1 UAUUAUUGGCUCACCGCUGUUAUUUUGGUGCUGAUAAAGAAUUGUGGGCAAUUC--AUCAGCAUUAUCAGGACAAAGAUUAGCGAGUC-----------AAUUAAUCAUCAGUUAAUACC ....((((((((.((..((((....((((((((((...((((((....))))))--))))))))))....))))..)....).)))))-----------))).................. ( -28.40) >DroYak_CAF1 3753 118 + 1 CAUUAUUGGCUCACCGCUGUUAUUUUGGUACCGAUAAAGAAUUGUGGCCAAUUC--AUUGGCAUUAUCAGAACAAAGACUAGCGAACCAAAUAAUGUUUAAUUAAUCAUCAGUUAAAUAC (((((((((.....(((((((.(((((((((((((...((((((....))))))--)))))...))))))))....))).))))..)).)))))))(((((((.......)))))))... ( -23.20) >consensus CAUUAUUGGCUCACCGCUGUUAUUUUGGUACCGAUAAAGAAUUGUGAGCAAUUC__AGCAGCAUUAUCAGAACAAAGAUUAGCGAGCCAAAUAAUGUUGAAUUAAUCAUCA_UCAAUUAC .....(((((((...((((((.(((((((((((((...((((((....))))))..)))))...))))))))....))).)))))))))).............................. (-18.19 = -18.75 + 0.56)


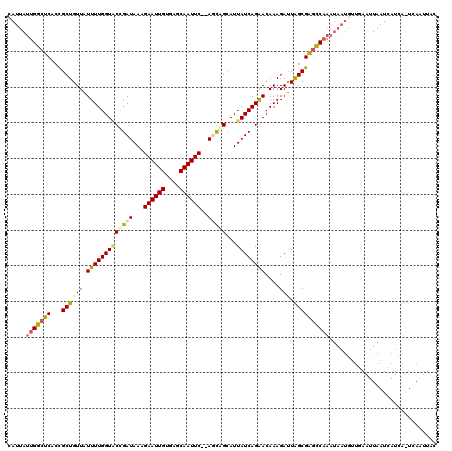
| Location | 3,953,413 – 3,953,530 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -18.58 |
| Energy contribution | -19.82 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3953413 117 - 22224390 GUAAGUGA-UGAUGAUUAAUUCAACAUUAUUUGGCCCGCCAAUCUUUGUUCUGAUAAUGCUGCU--GAAUUGCUCACAAUUCUUUAUCGGUACCAAAAUAACAGCAAUGAGCCAAUAAUG ..((((((-((.(((.....))).))))))))(((.........((((..(((((((.......--((((((....)))))).)))))))...))))....((....)).)))....... ( -23.40) >DroSim_CAF1 3862 119 - 1 GCAACUGA-UGAUGAUUAAUUCAACAUUAUUUGGCCCGCUAAUCUUUGUUCUGAUAAUUCUGCUGUGAAUUGCUCACAAUUCUUUAUCGGGACCAAAAUAACAGCGGUGAGCCAAUAAUU .....(((-((.(((.....))).))))).((((((((((....(((((((((((((......(((((.....))))).....))))))))).)))).....)))))...)))))..... ( -32.50) >DroEre_CAF1 3843 107 - 1 GGUAUUAACUGAUGAUUAAUU-----------GACUCGCUAAUCUUUGUCCUGAUAAUGCUGAU--GAAUUGCCCACAAUUCUUUAUCAGCACCAAAAUAACAGCGGUGAGCCAAUAAUA ..................(((-----------(.((((((.(((........)))..(((((((--((((((....))))))...))))))).............)))))).)))).... ( -23.70) >DroYak_CAF1 3753 118 - 1 GUAUUUAACUGAUGAUUAAUUAAACAUUAUUUGGUUCGCUAGUCUUUGUUCUGAUAAUGCCAAU--GAAUUGGCCACAAUUCUUUAUCGGUACCAAAAUAACAGCGGUGAGCCAAUAAUG ........................(((((((.((((((((.((..(((((.((....((((...--((((((....))))))......)))).)).)))))..))))))))))))))))) ( -27.50) >consensus GUAAUUAA_UGAUGAUUAAUUCAACAUUAUUUGGCCCGCUAAUCUUUGUUCUGAUAAUGCUGAU__GAAUUGCCCACAAUUCUUUAUCGGUACCAAAAUAACAGCGGUGAGCCAAUAAUG ..............................((((((((((.(((........)))..(((((((..((((((....))))))...))))))).............))))))))))..... (-18.58 = -19.82 + 1.25)

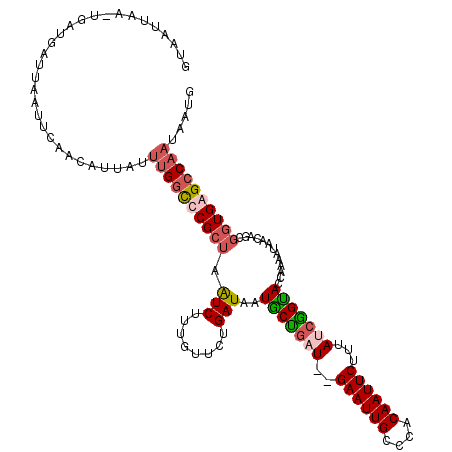

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:34 2006