
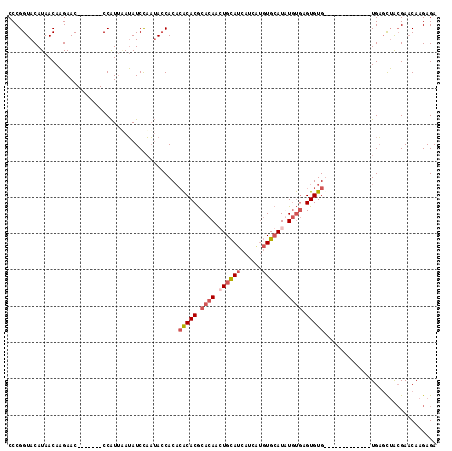
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,932,050 – 3,932,183 |
| Length | 133 |
| Max. P | 0.996785 |

| Location | 3,932,050 – 3,932,148 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.46 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -12.12 |
| Energy contribution | -13.75 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3932050 98 + 22224390 CCCGGUACAUAACAAGAAC-------CCAUUAAUAUCCAAUACCACACACACGCACAAGUGCAUCAUCAUGUGCAUGUGUGAGUGUG-------------UGAGCUACGAACAAGAGA ...(((.(.......).))-------)................(((((((.((((((.(..(((....)))..).)))))).)))))-------------))................ ( -26.80) >DroSec_CAF1 48563 98 + 1 CCCGGUACAUAACAAGAAC-------CCAUAAAUAUCCAAUACCACACACACGCACAAGUGCAUCAUCAUGUGCAUGUGUGAGUGUG-------------UGAGCUACGAACAAGAGA ...(((.(.......).))-------)................(((((((.((((((.(..(((....)))..).)))))).)))))-------------))................ ( -26.80) >DroYak_CAF1 53532 106 + 1 CCCGGUACAUAACAAGAAC-------CCAUUUAUAUCCAAUACCACACACACGCACAACUGCAUCAUCGUGUGCAUAUGUGAGUGUGUGUGUGUG-----UGAGCUACGAUCAAGAGA ...(((.(.......).))-------)............(((((((((((((.((((..(((((......)))))..)))).))))))))).)))-----)................. ( -29.90) >DroPer_CAF1 53423 106 + 1 CCCGGUACAUAACAUGAACGAUCGACCCGUUGAGAUCUCUAAGCAC--GCACACACAACUGUAUCCUC---UGAAUAUCCUUGUGUGU-------GUGUGUGCCUAGAGAACAAGAGA ..............(.((((.......)))).)(.((((((.((((--(((((((((...((((....---...)))).....)))))-------)))))))).)))))).)...... ( -33.20) >consensus CCCGGUACAUAACAAGAAC_______CCAUUAAUAUCCAAUACCACACACACGCACAACUGCAUCAUCAUGUGCAUAUGUGAGUGUG_____________UGAGCUACGAACAAGAGA ...............................................(((((.((((.((((((......)))))).)))).)))))............................... (-12.12 = -13.75 + 1.62)


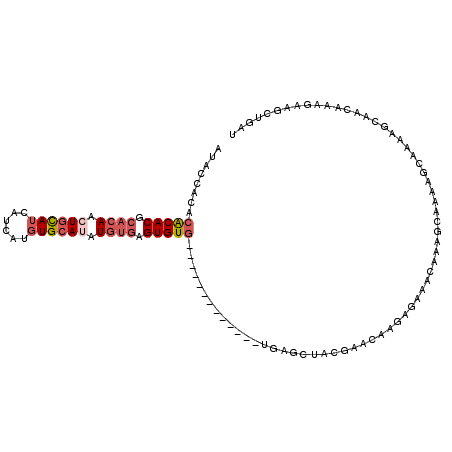
| Location | 3,932,082 – 3,932,183 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 72.33 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -12.12 |
| Energy contribution | -13.75 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3932082 101 + 22224390 AUACCACACACACGCACAAGUGCAUCAUCAUGUGCAUGUGUGAGUGUG-------------UGAGCUACGAACAAGAGAAACAAUGCAAAAGUAAUAGCAACAAAGAAGCUGAU ....(((((((.((((((.(..(((....)))..).)))))).)))))-------------))((((.(......)........(((..........))).......))))... ( -29.30) >DroSec_CAF1 48595 101 + 1 AUACCACACACACGCACAAGUGCAUCAUCAUGUGCAUGUGUGAGUGUG-------------UGAGCUACGAACAAGAGAAACAAAGCAAAAGCAAAAGCAACAAAGAAGCUGAU ....(((((((.((((((.(..(((....)))..).)))))).)))))-------------))((((.(......).........((....))..............))))... ( -29.40) >DroYak_CAF1 53564 108 + 1 AUACCACACACACGCACAACUGCAUCAUCGUGUGCAUAUGUGAGUGUGUGUGUGUG-----UGAGCUACGAUCAAGAGAA-CAAAGCAAACGCAAAAGCAACAAAGAAGCUGAU ....(((((((((.((((..(((((......)))))..)))).)))))))))((((-----(..(((..(.((....)).-)..)))..)))))..(((.........)))... ( -34.20) >DroPer_CAF1 53462 91 + 1 UAAGCAC--GCACACACAACUGUAUCCUC---UGAAUAUCCUUGUGUGU-------GUGUGUGCCUAGAGAACAAGAGAAACU-----------AAAGCAACCAAGAAGAUAAU ...((((--((((((((((..((((....---...))))..))))))))-------)))))).....................-----------.................... ( -23.00) >consensus AUACCACACACACGCACAACUGCAUCAUCAUGUGCAUAUGUGAGUGUG_____________UGAGCUACGAACAAGAGAAACAAAGCAAAAGCAAAAGCAACAAAGAAGCUGAU ........(((((.((((.((((((......)))))).)))).))))).................................................................. (-12.12 = -13.75 + 1.62)

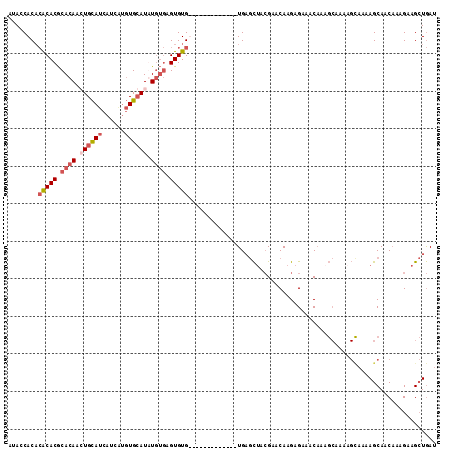
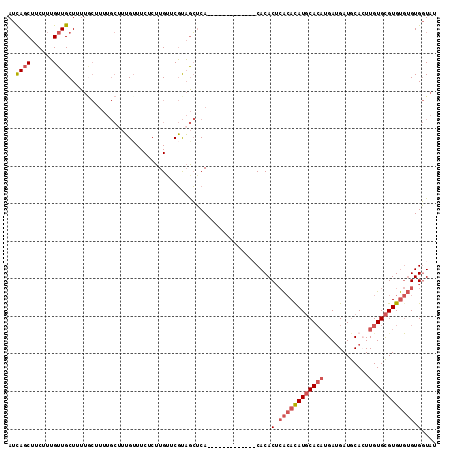
| Location | 3,932,082 – 3,932,183 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 72.33 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -13.11 |
| Energy contribution | -14.68 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3932082 101 - 22224390 AUCAGCUUCUUUGUUGCUAUUACUUUUGCAUUGUUUCUCUUGUUCGUAGCUCA-------------CACACUCACACAUGCACAUGAUGAUGCACUUGUGCGUGUGUGUGGUAU ..((((......)))).............................(((.(.((-------------(((((.((((..(((((.....).))))..)))).))))))).).))) ( -25.40) >DroSec_CAF1 48595 101 - 1 AUCAGCUUCUUUGUUGCUUUUGCUUUUGCUUUGUUUCUCUUGUUCGUAGCUCA-------------CACACUCACACAUGCACAUGAUGAUGCACUUGUGCGUGUGUGUGGUAU ..((((......)))).....((....))................(((.(.((-------------(((((.((((..(((((.....).))))..)))).))))))).).))) ( -26.40) >DroYak_CAF1 53564 108 - 1 AUCAGCUUCUUUGUUGCUUUUGCGUUUGCUUUG-UUCUCUUGAUCGUAGCUCA-----CACACACACACACUCACAUAUGCACACGAUGAUGCAGUUGUGCGUGUGUGUGGUAU ..((((......))))....(((((..(((.((-.((....)).)).)))..)-----)...(((((((((.((((..(((((.....).))))..)))).)))))))))))). ( -34.60) >DroPer_CAF1 53462 91 - 1 AUUAUCUUCUUGGUUGCUUU-----------AGUUUCUCUUGUUCUCUAGGCACACAC-------ACACACAAGGAUAUUCA---GAGGAUACAGUUGUGUGUGC--GUGCUUA ...........((..((...-----------.))..)).........(((((((.(((-------(((((....(.((((..---...)))))...)))))))).--))))))) ( -22.80) >consensus AUCAGCUUCUUUGUUGCUUUUGCUUUUGCUUUGUUUCUCUUGUUCGUAGCUCA_____________CACACUCACACAUGCACAUGAUGAUGCACUUGUGCGUGUGUGUGGUAU ..((((......))))......................................................(.((((((((((((............)))))))))))).).... (-13.11 = -14.68 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:28 2006