
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,908,771 – 3,908,883 |
| Length | 112 |
| Max. P | 0.555690 |

| Location | 3,908,771 – 3,908,883 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.07 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -17.19 |
| Energy contribution | -16.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3908771 112 + 22224390 UGCC-----AUAAAUUCGACGGGGACACAGAAAUCAAUGUAUUUCAUUAAUGGCCCGAAUGUGUGCCGGCUGUAAUGGGCUUUAUUGCC-AUUGUUGUUAUUAAUAUUGGCAUGCAAC .(((-----((.........(....)...(((((......)))))....))))).....(((((((((((.((((......)))).)))-..(((((....)))))..)))))))).. ( -29.40) >DroVir_CAF1 100738 100 + 1 CGCC-----------------CUAACACACAAAUCAAUGUAUUUCAUUAAUGGC-AAAAUGUGUGCCGGCUCUAAUAGGCUUUAUUGCCCAUUGUUAUUAUUAAUAUUGGCAUGCAAC .(((-----------------...........((.((((.....)))).)))))-....((((((((((........(((......)))...(((((....))))))))))))))).. ( -20.00) >DroPse_CAF1 35186 101 + 1 ---------A-GGAUCC------GACACAGAAAUCAAUGUAUUUCAUUAAUGGCUCAAAUGUGUGCCGGCUCUAAUAGGCUUUAUUGCC-AUUGUUGUUAUUAAUAUUGGCAUGCAAC ---------.-(((.((------(.(((.(((((......))))).......((......))))).))).))).....((.....((((-(.(((((....))))).))))).))... ( -23.40) >DroGri_CAF1 89828 110 + 1 CGCCCCACCU-GUGU------GUAACACAGAAAUCAAUGUAUUUCAUUAAUGGC-AAAAUGUGUGCCGGCUCUAAUAGGCUUUAUUGCCUAUUGUUAUUAUUAAUAUUGGCAUGCAAC .(((....((-(((.------....))))).....((((.....))))...)))-....((((((((((....(((((((......)))))))((((....)))).)))))))))).. ( -32.90) >DroWil_CAF1 15320 94 + 1 ---------------------GUAACACAAAAAUCAAUGUAUUUCAUUAAUGGU--AAAUGUGUGCCGGGUCUAAUGGGCUUUAUUGCC-AAUGUUCUUAUUAAUAUUGGCAUGCAAC ---------------------((((((((...(((((((.....)))...))))--...)))))(((.(......).))).....((((-((((((......)))))))))))))... ( -21.90) >DroPer_CAF1 22596 101 + 1 ---------A-GGAUCC------GACACAGAAAUCAAUGUAUUUCAUUAAUGGCUCAAAUGUGUGCCGGCUCUAAUAGGCUUUAUUGCC-AUUGUUGUUAUUAAUAUUGGCAUGCAAC ---------.-(((.((------(.(((.(((((......))))).......((......))))).))).))).....((.....((((-(.(((((....))))).))))).))... ( -23.40) >consensus _________A_G_AU______GUAACACAGAAAUCAAUGUAUUUCAUUAAUGGC_CAAAUGUGUGCCGGCUCUAAUAGGCUUUAUUGCC_AUUGUUGUUAUUAAUAUUGGCAUGCAAC ...................................((((.....))))...........((((((((((...((((((((......))).........)))))...)))))))))).. (-17.19 = -16.97 + -0.22)

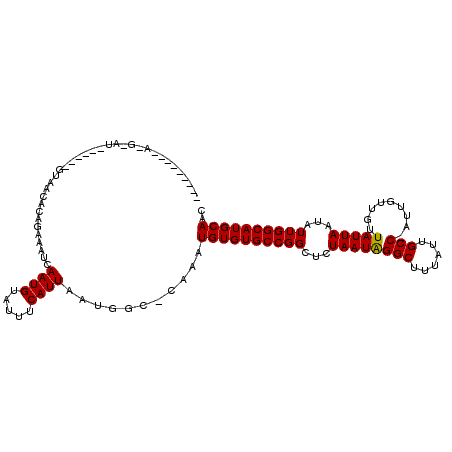
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:19 2006