
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,887,075 – 3,887,184 |
| Length | 109 |
| Max. P | 0.743014 |

| Location | 3,887,075 – 3,887,184 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -43.80 |
| Consensus MFE | -33.23 |
| Energy contribution | -34.73 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
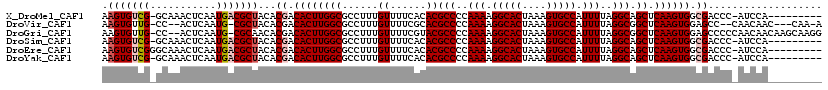
>X_DroMel_CAF1 3887075 109 + 22224390 ---------UGGAU-GGGUCGCCACUUGAGCUGCCUAAAAUGGCACUUUAGUGCCUUUUGGGGCGUGUGAAAACAAAGGCGCCAAGUGUCGUGUAGCGUCAUUGAGUUUGC-CGACACUU ---------.....-.((.((((......((..(((((((.(((((....)))))))))))))).(((....)))..))))))((((((((.(((((........).))))-)))))))) ( -43.10) >DroVir_CAF1 70829 110 + 1 U-UUG---GUUGUUG--GGCUCCACUUGAGCCGCCUAAAAUGGCACUUUAGUGCCUUUUGGGGCGUGCGAAAACAAAGGCGCCAAGUGUCGUGUAGCG-CAUUGAGU--GG-CAACACUU .-(..---(((((((--.((..((((((.(((((((.(((.(((((....))))).))).))))((......))...)))..))))))..)).)))))-.))..)((--(.-...))).. ( -45.40) >DroGri_CAF1 59357 116 + 1 CCUUGCUUGUUGUUGGGGGCUCCACUUGAGCCGCCUAAAAUGGCACUUUAGUGCCUUUUGGGGCGUACGAAAACAAAGGCGCCAAGUGUCGUGUUGCG-CAUUGAGU--GG-CAACACUU (((.((.....)).))).....((((((.(((((((.(((.(((((....))))).))).))))((......))...)))..))))))..(((((((.-(((...))--))-)))))).. ( -45.00) >DroSim_CAF1 1714 109 + 1 ---------UGGAU-GGGUCGCCACUUGAGCUGCCUAAAAUGGCACUUUAGUGCCUUUUGGGGCGUGUGAAAACAAAGGCGCCAAGUGUCGUGUAGCGUCAUUGAGUUUGC-CGACACUU ---------.....-.((.((((......((..(((((((.(((((....)))))))))))))).(((....)))..))))))((((((((.(((((........).))))-)))))))) ( -43.10) >DroEre_CAF1 5300 110 + 1 ---------UGGAU-GGGUCGCCACUUGAGCUGCCUAAAAUGGCACUUUAGUGCCUUUUGGGGCGUGUGAAAACAAAGGCGCCAAGUGUCGUGUAGCGUCAUUGAGUUUGCCCGACACUU ---------.....-.((.((((......((..(((((((.(((((....)))))))))))))).(((....)))..))))))((((((((.(((((........).)))).)))))))) ( -43.10) >DroYak_CAF1 7109 109 + 1 ---------UGGAU-GGGUCGCCACUUGAGCUGCCUAAAAUGGCACUUUAGUGCCUUUUGGGGCGUGUGAAAACAAAGGCGCCAAGUGUCGUGUAGCGUCAUUGAGUUUGC-CGACACUU ---------.....-.((.((((......((..(((((((.(((((....)))))))))))))).(((....)))..))))))((((((((.(((((........).))))-)))))))) ( -43.10) >consensus _________UGGAU_GGGUCGCCACUUGAGCUGCCUAAAAUGGCACUUUAGUGCCUUUUGGGGCGUGUGAAAACAAAGGCGCCAAGUGUCGUGUAGCGUCAUUGAGUUUGC_CGACACUU ................((.((((......((..(((((((.(((((....))))))))))))))((......))...))))))((((((((.(((((........)).))).)))))))) (-33.23 = -34.73 + 1.50)


| Location | 3,887,075 – 3,887,184 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -28.00 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.743014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
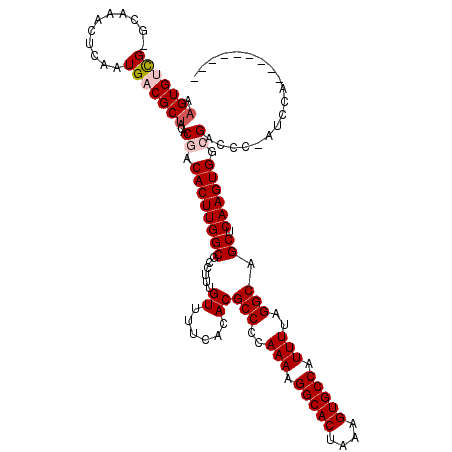
>X_DroMel_CAF1 3887075 109 - 22224390 AAGUGUCG-GCAAACUCAAUGACGCUACACGACACUUGGCGCCUUUGUUUUCACACGCCCCAAAAGGCACUAAAGUGCCAUUUUAGGCAGCUCAAGUGGCGACCC-AUCCA--------- .(((((((-..........)))))))...((.((((((((.....(((....))).(((..(((.(((((....))))).)))..))).)).)))))).))....-.....--------- ( -33.70) >DroVir_CAF1 70829 110 - 1 AAGUGUUG-CC--ACUCAAUG-CGCUACACGACACUUGGCGCCUUUGUUUUCGCACGCCCCAAAAGGCACUAAAGUGCCAUUUUAGGCGGCUCAAGUGGAGCC--CAACAAC---CAA-A ...(((((-((--((((((.(-(((((.........))))))..))).....((.((((..(((.(((((....))))).)))..))))))...)))))....--)))))..---...-. ( -36.30) >DroGri_CAF1 59357 116 - 1 AAGUGUUG-CC--ACUCAAUG-CGCAACACGACACUUGGCGCCUUUGUUUUCGUACGCCCCAAAAGGCACUAAAGUGCCAUUUUAGGCGGCUCAAGUGGAGCCCCCAACAACAAGCAAGG ..((((((-((--(.....))-.)))))))..(((((((.(((...((......))(((..(((.(((((....))))).)))..)))))))))))))...................... ( -39.40) >DroSim_CAF1 1714 109 - 1 AAGUGUCG-GCAAACUCAAUGACGCUACACGACACUUGGCGCCUUUGUUUUCACACGCCCCAAAAGGCACUAAAGUGCCAUUUUAGGCAGCUCAAGUGGCGACCC-AUCCA--------- .(((((((-..........)))))))...((.((((((((.....(((....))).(((..(((.(((((....))))).)))..))).)).)))))).))....-.....--------- ( -33.70) >DroEre_CAF1 5300 110 - 1 AAGUGUCGGGCAAACUCAAUGACGCUACACGACACUUGGCGCCUUUGUUUUCACACGCCCCAAAAGGCACUAAAGUGCCAUUUUAGGCAGCUCAAGUGGCGACCC-AUCCA--------- (((((((((((............)))...))))))))((((((.............(((..(((.(((((....))))).)))..))).((....)))))).)).-.....--------- ( -34.20) >DroYak_CAF1 7109 109 - 1 AAGUGUCG-GCAAACUCAAUGACGCUACACGACACUUGGCGCCUUUGUUUUCACACGCCCCAAAAGGCACUAAAGUGCCAUUUUAGGCAGCUCAAGUGGCGACCC-AUCCA--------- .(((((((-..........)))))))...((.((((((((.....(((....))).(((..(((.(((((....))))).)))..))).)).)))))).))....-.....--------- ( -33.70) >consensus AAGUGUCG_GCAAACUCAAUGACGCUACACGACACUUGGCGCCUUUGUUUUCACACGCCCCAAAAGGCACUAAAGUGCCAUUUUAGGCAGCUCAAGUGGCGACCC_AUCCA_________ .(((((((...........)))))))...((.((((((((......((......))(((..(((.(((((....))))).)))..))).)).)))))).))................... (-28.00 = -28.62 + 0.61)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:06:07 2006