| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,860,465 – 3,860,586 |
| Length | 121 |
| Max. P | 0.894430 |

| Location | 3,860,465 – 3,860,566 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -18.07 |
| Energy contribution | -17.93 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
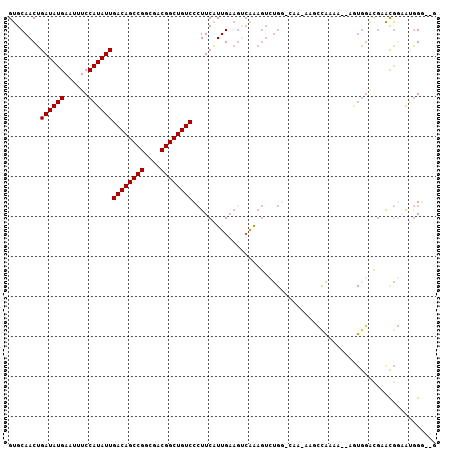
>X_DroMel_CAF1 3860465 101 + 22224390 C---CCAUACCGUUCGUCCACU--UUUUGGCUUUUUG-GCAGACUUUGACUUCAAUGAAGGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCAC .---(((((((.((((((((..--...))(((....)-)).))).(((....))).)))))((((((((....))))))))..)))))................... ( -26.70) >DroPse_CAF1 47634 89 + 1 CCUCCUCUGGUGUCC-UCC----------CCUG-CUGUC------CUGGCUUCAACGAAUGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCAC ......(((((((..-(((----------...(-((...------..)))...........((((((((....)))))))).....))).....)))))))...... ( -23.80) >DroSec_CAF1 43594 101 + 1 C--CCCAUUCCGUGCGUCCACU--UUUUGGCUU-UUG-GCAGACUUUGACUUCAAUGAAGGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCAC .--........(((((((....--.((((.(..-..)-.))))....))).....(((...((((((((....))))))))..(((((....))))))))...)))) ( -27.30) >DroEre_CAF1 42442 96 + 1 C-CCCCAUUCCGUUCGACCAUU--UUUUGG--------CCAGACUUUGACUUCAAUGAAGGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCAC .-.....((((((..(.(((..--...)))--------.)..)).....((((...)))).((((((((....)))))))).....))))................. ( -25.00) >DroYak_CAF1 38568 96 + 1 C---CCACUCCGUUCGCCCACUGGUUUUGG--------CCAGACUUUGACUUCAAUGAAGGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCAC .---....(((((....((.(((((....)--------)))).(.(((....))).)..))((((((((....))))))))...))))).................. ( -27.90) >DroAna_CAF1 44564 98 + 1 C--CCCCUCACACAAAUAUAUU--UUUCAUUUU-UUGGCCAGACUUC----ACAAUGAAGGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCAC .--........((..((((..(--((((((...-.........((((----.....)))).((((((((....))))))))...)))))))...))))..))..... ( -22.40) >consensus C__CCCAUUCCGUUCGUCCACU__UUUUGGCUU_UUG_CCAGACUUUGACUUCAAUGAAGGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCAC ....................................................((((.....((((((((....)))))))).((((((....))))))..))))... (-18.07 = -17.93 + -0.14)



| Location | 3,860,465 – 3,860,566 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -20.40 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3860465 101 - 22224390 GUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUUGAAGUCAAAGUCUGC-CAAAAAGCCAAAA--AGUGGACGAACGGUAUGG---G ...................(((((..((((((((....))))))))(((((.(((....))).(((..(-.............--.)..)))))).)))))))---. ( -30.24) >DroPse_CAF1 47634 89 - 1 GUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCAUUCGUUGAAGCCAG------GACAG-CAGG----------GGA-GGACACCAGAGGAGG (((((((.((((((......))))))((((((((....)))))))).....))))...((..------(....-)..)----------)..-...)))......... ( -26.40) >DroSec_CAF1 43594 101 - 1 GUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUUGAAGUCAAAGUCUGC-CAA-AAGCCAAAA--AGUGGACGCACGGAAUGGG--G ((((.(((((((((......))))))((((((((....))))))))..........)))....(((..(-...-.........--.)..)))))))........--. ( -32.42) >DroEre_CAF1 42442 96 - 1 GUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUUGAAGUCAAAGUCUGG--------CCAAAA--AAUGGUCGAACGGAAUGGGG-G ........((((((......))))))((((((((....))))))))(((((((((....)...((.(((--------(((...--..)))))).))..)))))))-) ( -34.30) >DroYak_CAF1 38568 96 - 1 GUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUUGAAGUCAAAGUCUGG--------CCAAAACCAGUGGGCGAACGGAGUGG---G ........((((((......))))))((((((((....))))))))(((((......(((.....((((--------......))))..))).....))).))---. ( -31.00) >DroAna_CAF1 44564 98 - 1 GUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUUGU----GAAGUCUGGCCAA-AAAAUGAAA--AAUAUAUUUGUGUGAGGGG--G ...(..((.(((..(((....(((((((((((((....))))))))..((((((.(----(.........)).-..)))))).--))))))))..))).))..)--. ( -28.30) >consensus GUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUUGAAGUCAAAGUCUGG_CAA_AAGCCAAAA__AGUGGACGAACGGAAUGGG__G ........((((((......))))))((((((((....))))))))............................................................. (-20.40 = -20.40 + 0.00)


| Location | 3,860,496 – 3,860,586 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -21.12 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
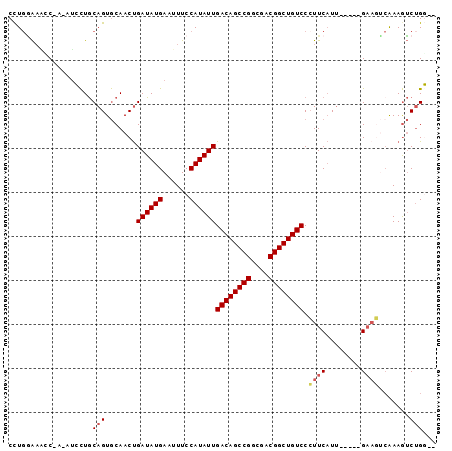
>X_DroMel_CAF1 3860496 90 - 22224390 CUUGGAAGUC-AACUGCUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUU-----GAAGUCAAAGUCUGC-C (((.((..((-((.(((......)))...((((((......))))))((((((((....))))))))......))-----))..)).))).....-. ( -28.20) >DroSec_CAF1 43625 89 - 1 CCUGGAAGGC-A-GUGCUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUU-----GAAGUCAAAGUCUGC-C ...((.((((-.-.((((.(((((.....((((((......))))))((((((((....))))))))....))))-----).)).))..)))).)-) ( -32.70) >DroEre_CAF1 42469 88 - 1 CCUGAAAACC-A-GUCCUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUU-----GAAGUCAAAGUCUGG-- ........((-(-(.(((.(((((.....((((((......))))))((((((((....))))))))....))))-----).)).....).))))-- ( -28.20) >DroWil_CAF1 42044 93 - 1 CCUGUGUUCC-U-AGCUUACAGUACAACCGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCGUUCCUAAGGGAGAACGCAAAAGCGGG-- ..((((((((-(-..((((....((....((((((......))))))((((((((....)))))))).))...)))))).)))))))........-- ( -31.20) >DroYak_CAF1 38595 88 - 1 CCUGGAAAGC-A-AUCCUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUU-----GAAGUCAAAGUCUGG-- .(..((....-.-...((.(((((.....((((((......))))))((((((((....))))))))....))))-----).))......))..)-- ( -26.94) >DroAna_CAF1 44595 87 - 1 CCUCGAACUCGA-AUCCUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUU-----GU----GAAGUCUGGCC (((((....)))-.....((((((.....((((((......))))))((((((((....))))))))....))))-----))----.......)).. ( -27.40) >consensus CCUGGAAACC_A_AUCCUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCCUUCAUU_____GAAGUCAAAGUCUGG__ ...................(((.......((((((......))))))((((((((....)))))))).((((........)))).......)))... (-21.12 = -21.57 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:55 2006