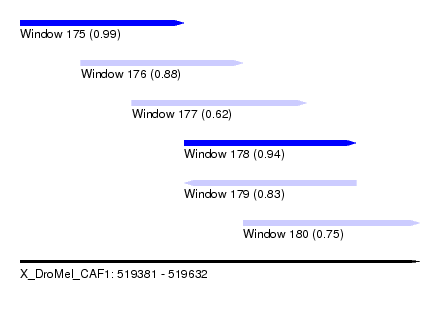
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 519,381 – 519,632 |
| Length | 251 |
| Max. P | 0.989568 |

| Location | 519,381 – 519,484 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.73 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -4.39 |
| Energy contribution | -4.23 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.17 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 519381 103 + 22224390 AGAAGCGGGAGCAGGAGCAGGAGCAGGAGCAGGAGCU--UCGAAAG-AUGCGGG-C-AUC-CAG-AAGG--UCGCAUAA-AAGUGUGGC-AU--AAAAUAUGAAUU--AAAAAC--CAGU ....((....)).((.((....)).(((((....((.--((....)-).))..)-)-.))-)..-...(--((((((..-..)))))))-..--............--.....)--)... ( -27.20) >DroSec_CAF1 5683 91 + 1 AGGAGCGGGUGCA------------GGGGCAGGAGCU--UCGAAAG-AUGCGGG-C-AGC-CAG-AAGG--UCGCAUAA-GCUUGUGGC-AU--AAAAUAUGAAAU--AAAAAG--CAGG ....((((.(((.------------.........((.--((....)-).))..)-)-).)-)..-...(--(((((...-...))))))-..--............--.....)--)... ( -22.60) >DroSim_CAF1 6059 91 + 1 AGGAGCGGGUGCA------------GGGGCAGGAGCU--UCGAAAG-AUGCGGG-C-AGC-CAG-AAGG--UCGCAUAA-AAGUGUGGC-AU--AAAAUAUGAAUU--AAAAAG--CAGG ....((((.(((.------------.........((.--((....)-).))..)-)-).)-)..-...(--((((((..-..)))))))-..--............--.....)--)... ( -23.20) >DroEre_CAF1 5483 89 + 1 --GACCAGGAGCA------------GGAGCAGGGGCC--UCGAAAG-AUGCGGG-C-AGC-CAG-AAGG--UCGCAUAA-AAGUGUGCC-AU--GGAAUAUGAAUU--AAAAAC--CAGG --((((....((.------------...)).((.(((--((....)-)....))-)-..)-)..-..))--))(((((.-...))))).-.(--((..........--.....)--)).. ( -20.96) >DroYak_CAF1 5797 108 + 1 GGGGGAAGGACCA------------GGAGCAGGGUACUCUCGUAAGAAUGCGGGGCAAGCCCAGGAAGGGUCCGCAUAAAAAGAGUGCCCAUUAGAAAUAUGAUAUAUAAAAACCCCAGG ((((.........------------......((((((((((....))((((((.....((((.....)))))))))).....))))))))((((......)))).........))))... ( -38.60) >consensus AGGAGCGGGAGCA____________GGAGCAGGAGCU__UCGAAAG_AUGCGGG_C_AGC_CAG_AAGG__UCGCAUAA_AAGUGUGGC_AU__AAAAUAUGAAUU__AAAAAC__CAGG ...............................................((((((..................))))))........................................... ( -4.39 = -4.23 + -0.16)

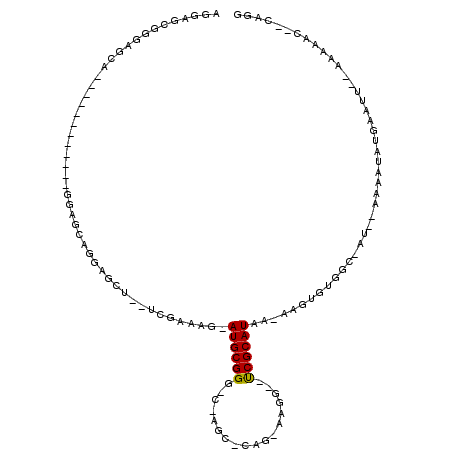
| Location | 519,419 – 519,521 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -16.60 |
| Energy contribution | -19.10 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 519419 102 + 22224390 CGAAAGAUGCGGGCAUCCAGAAGGUCGCAUAAAAGUGUGGCAUAAAAUAUGAAUUAAAAACCAGUAGGAAAGGGUGGUAAAUGCCGCGGGGGUCCGAGGGUG--- (....)...(((((..((.....(((((((....)))))))...................((....)).....(((((....)))))))..)))))......--- ( -27.70) >DroSec_CAF1 5709 102 + 1 CGAAAGAUGCGGGCAGCCAGAAGGUCGCAUAAGCUUGUGGCAUAAAAUAUGAAAUAAAAAGCAGGAGGAAAGGGCGGUAAAUGCCGCGGGGGUCCGAGGGUG--- (....)...(((((..((.....((((((......))))))................................(((((....)))))))..)))))......--- ( -28.00) >DroSim_CAF1 6085 102 + 1 CGAAAGAUGCGGGCAGCCAGAAGGUCGCAUAAAAGUGUGGCAUAAAAUAUGAAUUAAAAAGCAGGAGGAAAGGGCGGUAAAUGCCGCGGGGGUCCGAGGGUG--- (....)...(((((..((.....(((((((....)))))))................................(((((....)))))))..)))))......--- ( -28.60) >DroEre_CAF1 5507 87 + 1 CGAAAGAUGCGGGCAGCCAGAAGGUCGCAUAAAAGUGUGCCAUGGAAUAUGAAUUAAAAACCAGGAGGACAGGGUG------------------CGAGGGUGGGA (....)((.(..(((.((....((.(((((....)))))))...................((....))...)).))------------------)..).)).... ( -18.70) >consensus CGAAAGAUGCGGGCAGCCAGAAGGUCGCAUAAAAGUGUGGCAUAAAAUAUGAAUUAAAAACCAGGAGGAAAGGGCGGUAAAUGCCGCGGGGGUCCGAGGGUG___ (....)...(((((..((.....(((((((....)))))))................................(((((....)))))))..)))))......... (-16.60 = -19.10 + 2.50)

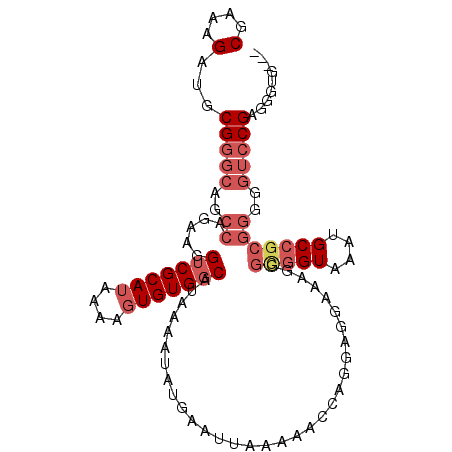
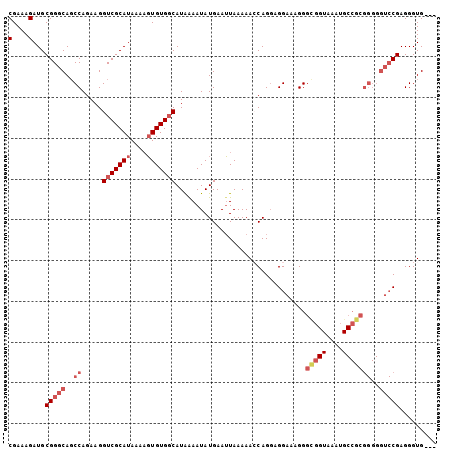
| Location | 519,451 – 519,561 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.71 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -17.05 |
| Energy contribution | -18.55 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 519451 110 + 22224390 AAGUGUGGCAUAAAAUAUGAAUUAAAAACCAGUAGGAAAGGGUGGUAAAUGCCGCGGGGGUCCGAGGGUG---CAGGAUGAUGGACAAGCCGGAAAACCGGCAGGAAACUUCA ....(((((((..(((....)))....((((.(.......).))))..)))))))(((..(((....((.---((......)).))..(((((....))))).)))..))).. ( -31.10) >DroSec_CAF1 5741 110 + 1 GCUUGUGGCAUAAAAUAUGAAAUAAAAAGCAGGAGGAAAGGGCGGUAAAUGCCGCGGGGGUCCGAGGGUG---CAGGAUGAUGGACAAGCCGGAAACCCGGCAGGAAACUUCG ((((((..((((...))))..)))...))).((((......(((((....)))))(((..((((...((.---((......)).))....))))..))).........)))). ( -33.40) >DroSim_CAF1 6117 110 + 1 AAGUGUGGCAUAAAAUAUGAAUUAAAAAGCAGGAGGAAAGGGCGGUAAAUGCCGCGGGGGUCCGAGGGUG---CAGGAUGAUGGACAAGCCGGAAACCCGGCAGGAAACUUCG ..((((........))))..........((...........(((((....)))))(((..((((...((.---((......)).))....))))..))).))((....))... ( -29.70) >DroEre_CAF1 5539 94 + 1 AAGUGUGCCAUGGAAUAUGAAUUAAAAACCAGGAGGACAGGGUG------------------CGAGGGUGGGACAGGAUGAUGGACAAGCCGGAAAG-CGGCAGGAAACUUCG ....(..((.((.....((..........))......)).))..------------------)((((...(..((......))..)..((((.....-))))......)))). ( -19.60) >consensus AAGUGUGGCAUAAAAUAUGAAUUAAAAACCAGGAGGAAAGGGCGGUAAAUGCCGCGGGGGUCCGAGGGUG___CAGGAUGAUGGACAAGCCGGAAACCCGGCAGGAAACUUCG ..................................(((....(((((....))))).....)))((((......((......)).....(((((....)))))......)))). (-17.05 = -18.55 + 1.50)

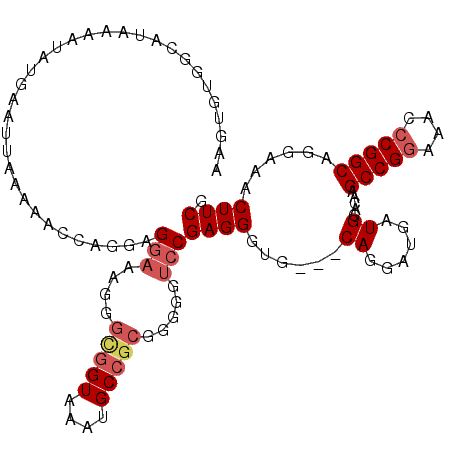
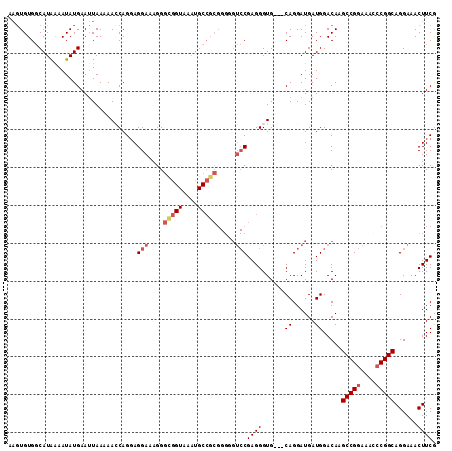
| Location | 519,484 – 519,592 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -45.17 |
| Consensus MFE | -42.42 |
| Energy contribution | -42.53 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 519484 108 + 22224390 AGGAAAGGGUGGUAAAUGCCGCGGGGGUCCGAGGGUGCAGGAUGAUGGACAAGCCGGAAAACCGGCAGGAAACUUCAUGACCGUUCGAGUUCCUGCCCGGCUUGCAUA ........(((((....)))))(.(((((....((.((((((...(((((..(((((....)))))((....))........)))))...))))))))))))).)... ( -41.10) >DroSec_CAF1 5774 108 + 1 AGGAAAGGGCGGUAAAUGCCGCGGGGGUCCGAGGGUGCAGGAUGAUGGACAAGCCGGAAACCCGGCAGGAAACUUCGUGACCGUUUGAGUUCCUGCCGGGCUUGCACA .(((....(((((....))))).....)))....(((((((....(((.....)))....(((((((((((...(((........))).)))))))))))))))))). ( -47.20) >DroSim_CAF1 6150 108 + 1 AGGAAAGGGCGGUAAAUGCCGCGGGGGUCCGAGGGUGCAGGAUGAUGGACAAGCCGGAAACCCGGCAGGAAACUUCGUGACCGUUUGAGUUCCUGCCGGGCUUGCACA .(((....(((((....))))).....)))....(((((((....(((.....)))....(((((((((((...(((........))).)))))))))))))))))). ( -47.20) >consensus AGGAAAGGGCGGUAAAUGCCGCGGGGGUCCGAGGGUGCAGGAUGAUGGACAAGCCGGAAACCCGGCAGGAAACUUCGUGACCGUUUGAGUUCCUGCCGGGCUUGCACA ........(((((....)))))(.(((((((.....((((((.(((((.((.(((((....)))))((....))...)).))))).....))))))))))))).)... (-42.42 = -42.53 + 0.11)

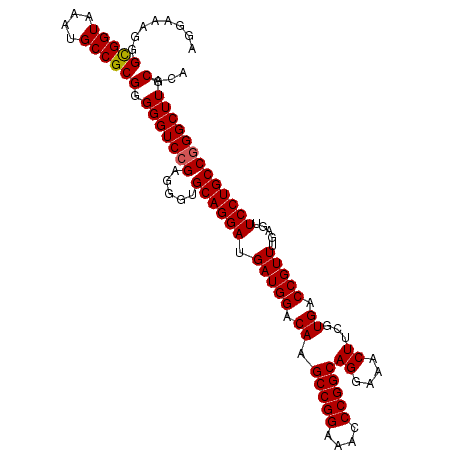
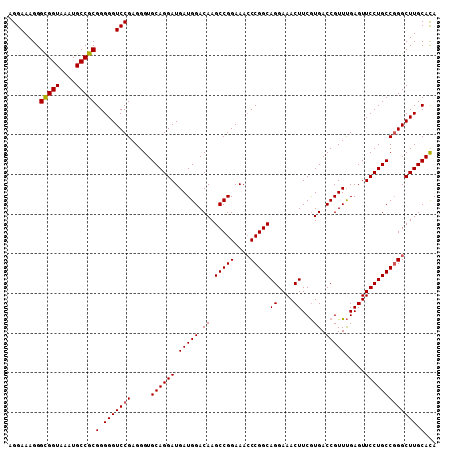
| Location | 519,484 – 519,592 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.30 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -32.59 |
| Energy contribution | -33.03 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 519484 108 - 22224390 UAUGCAAGCCGGGCAGGAACUCGAACGGUCAUGAAGUUUCCUGCCGGUUUUCCGGCUUGUCCAUCAUCCUGCACCCUCGGACCCCCGCGGCAUUUACCACCCUUUCCU ...(((((((.((((((((((((........)).)).))))))))((....)))))))))..............((.(((....))).)).................. ( -32.20) >DroSec_CAF1 5774 108 - 1 UGUGCAAGCCCGGCAGGAACUCAAACGGUCACGAAGUUUCCUGCCGGGUUUCCGGCUUGUCCAUCAUCCUGCACCCUCGGACCCCCGCGGCAUUUACCGCCCUUUCCU .((((((((((((((((((((....((....)).)).))))))))))))))..((..((.....)).)).))))....(((.....((((......))))....))). ( -40.80) >DroSim_CAF1 6150 108 - 1 UGUGCAAGCCCGGCAGGAACUCAAACGGUCACGAAGUUUCCUGCCGGGUUUCCGGCUUGUCCAUCAUCCUGCACCCUCGGACCCCCGCGGCAUUUACCGCCCUUUCCU .((((((((((((((((((((....((....)).)).))))))))))))))..((..((.....)).)).))))....(((.....((((......))))....))). ( -40.80) >consensus UGUGCAAGCCCGGCAGGAACUCAAACGGUCACGAAGUUUCCUGCCGGGUUUCCGGCUUGUCCAUCAUCCUGCACCCUCGGACCCCCGCGGCAUUUACCGCCCUUUCCU ((.(((((((.((((((((((....((....)).)).))))))))((....))))))))).))...............(((.....((((......))))....))). (-32.59 = -33.03 + 0.45)

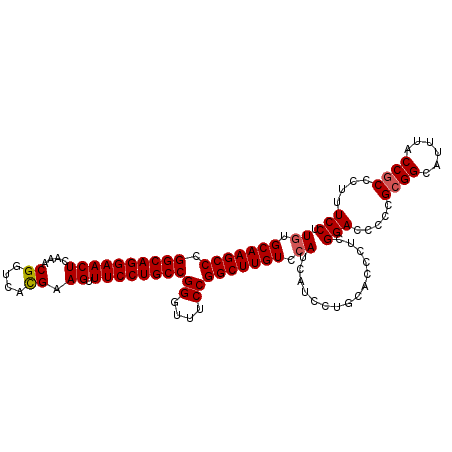
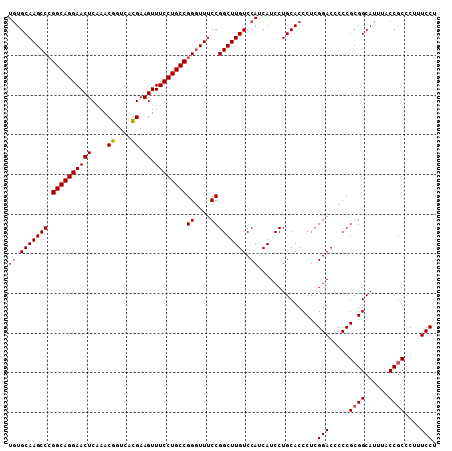
| Location | 519,521 – 519,632 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -37.76 |
| Consensus MFE | -30.21 |
| Energy contribution | -30.84 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 519521 111 + 22224390 CAGGAUGAUGGACAAGCCGGAAAACCGGCAGGAAACUUCAUGACCGUUCGAGUUCCUGCCC---------GGCUUGCAUAGACAAAAGUAAAAAGUGAUAAGCGGCUUAGGACUCGGCUA ..((.(.(((((...(((((....))))).(....)))))).)))..((((((((..(((.---------.((((((((..((....)).....))).))))))))...))))))))... ( -36.10) >DroSec_CAF1 5811 111 + 1 CAGGAUGAUGGACAAGCCGGAAACCCGGCAGGAAACUUCGUGACCGUUUGAGUUCCUGCCG---------GGCUUGCACAGACAAAAGUGAAAAGUGAUAAGCGGCUUAGGACUCGGCUA ..((.(((.(..(((((((....)((((((((((...(((........))).)))))))))---------)((((((((...............))).)))))))))).)..)))).)). ( -39.86) >DroSim_CAF1 6187 111 + 1 CAGGAUGAUGGACAAGCCGGAAACCCGGCAGGAAACUUCGUGACCGUUUGAGUUCCUGCCG---------GGCUUGCACAGACAAAAGUGAAAAGUGAUAAGCGGCUUAGGACUCGGCUA ..((.(((.(..(((((((....)((((((((((...(((........))).)))))))))---------)((((((((...............))).)))))))))).)..)))).)). ( -39.86) >DroEre_CAF1 5594 119 + 1 CAGGAUGAUGGACAAGCCGGAAAG-CGGCAGGAAACUUCGUGACCGUUCGAGUUCCUGACGGGAACUCCUGGCUGGCAUAGACAAAAGUGAAACGUGAUAAGCGGCUUAGGACUCGGCUA ..((.(((.(..(.((((((..((-(((((.((....)).)).))))).(((((((.....)))))))))))))(((....((....))....(((.....))))))..)..)))).)). ( -35.20) >consensus CAGGAUGAUGGACAAGCCGGAAACCCGGCAGGAAACUUCGUGACCGUUCGAGUUCCUGCCG_________GGCUUGCACAGACAAAAGUGAAAAGUGAUAAGCGGCUUAGGACUCGGCUA ..((.(((.(..((((((.......(((((((((...(((........))).)))))))))..........((((((((...............))).)))))))))).)..)))).)). (-30.21 = -30.84 + 0.63)

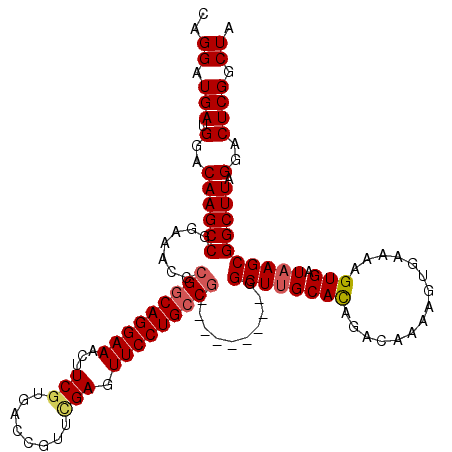
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:09 2006