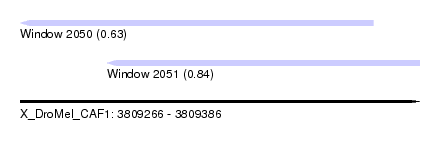
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,809,266 – 3,809,386 |
| Length | 120 |
| Max. P | 0.837944 |

| Location | 3,809,266 – 3,809,372 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.66 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -16.61 |
| Energy contribution | -16.67 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
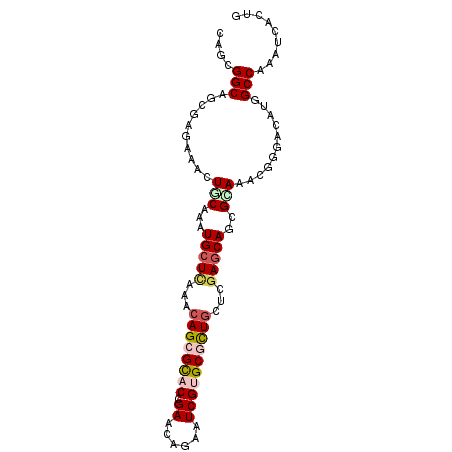
>X_DroMel_CAF1 3809266 106 - 22224390 AGCUUCUGAUGCUCACACAGCGCACGGAACAAAAUCGUGCUUUGUUGGAGCAGCGCAAGAGGGAUCUGGCCAAGUCUCUUCUUUCUGUAAAGUCGAAUA--UUGGCCA--- ..((((((.(((((..((((.((((((.......)))))).))))..)))))...).))))).....((((((...((..((((....))))..))...--)))))).--- ( -35.80) >DroVir_CAF1 10291 109 - 1 AGCUGCAAAUGCUUAAUCAGCGCACCGAACAGAAUCGUGCGCUGCUCGAGCAGCGCAAACGUGACAUGGCCAAGUCGCUGCAGUCCGUCAAGACUGGCC--UCCGCCAGAG .((((((..((((..(.((((((((.((......)))))))))).)..))))........(((((........))))))))))).........(((((.--...))))).. ( -42.80) >DroGri_CAF1 10230 109 - 1 AACUGCAAAUGCUCAAUCAGCGCACCGAGCAGAAUCGUGCGCUGCUCGAGCAGCGUAAACGGGACAUGGCCAAAUCACUACAGUCCGUCAAAUCGGGUC--UGAGGCAGAG ..((((...(((((...((((((((.((......))))))))))...))))).........((......))......((.(((((((......)))).)--)))))))).. ( -38.50) >DroEre_CAF1 8149 106 - 1 AGCUUCUGAUGCUCACACAGCGCACGGAACAAAAUCGUGCUUUGUUAGAACAGCGUAAAAGAGAUCUGGCCAAGUCUCUUCUUUCUGUAAAGUCGAAUA--UUGGCCA--- .((((((((((((.....)))((((((.......))))))...))))))..))).............((((((...((..((((....))))..))...--)))))).--- ( -29.00) >DroMoj_CAF1 6377 109 - 1 AGUUGCAAAUGCUGAACCAGCGCACCGAGCAAAAUCGUGCGCUCCUCGAGCAGCGCAAACGCGACAUGGCCAAAUCGCUGCAGUCGGUGAAAUCGGGCA--UGCGGCAGAG .((((((..((((((((((((((((.((......)))))))))......((((((.....((......)).....))))))....)))....)).))))--)))))).... ( -38.50) >DroAna_CAF1 8610 107 - 1 AACUACAGAUGCUCACCCAGCGUACGGAGCAGAAUCGCGCUCUGCUGGAGCAGCGGAAGCGGGAUCUGGCCAAAUCCCUGCUCUCGGUCAAGACCAGCUCCAUGUCC---- ........(((((.....))))).((((((........)))))).((((((...(..((((((((........))))).)))..)(((....))).)))))).....---- ( -37.40) >consensus AGCUGCAAAUGCUCAAACAGCGCACCGAACAAAAUCGUGCGCUGCUCGAGCAGCGCAAACGGGACAUGGCCAAAUCGCUGCAGUCCGUCAAGUCGAGCA__UUGGCCA___ .........(((((...(((.((((.((......)))))).)))...)))))........(((((........)))))................................. (-16.61 = -16.67 + 0.06)



| Location | 3,809,292 – 3,809,386 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3809292 94 - 22224390 CAGUGGCAGCGAGAAGCUUCUGAUGCUCACACAGCGCACGGAACAAAAUCGUGCUUUGUUGGAGCAGCGCAAGAGGGAUCUGGCCAAGUCUCUU .((.(((.((.(((..((((((.(((((..((((.((((((.......)))))).))))..)))))...).)))))..))).))...))).)). ( -32.30) >DroVir_CAF1 10320 94 - 1 CAGCGGCAGCGAGAAGCUGCAAAUGCUUAAUCAGCGCACCGAACAGAAUCGUGCGCUGCUCGAGCAGCGCAAACGUGACAUGGCCAAGUCGCUG ..((((((((.....)))))...((((..(.((((((((.((......)))))))))).)..)))).)))....(((((........))))).. ( -41.00) >DroPse_CAF1 7813 94 - 1 UAGUGGCAGUGAAAAAGUUCUGAUGCUCACACAGCGCACGGAACAGAAUCGGGCGCUGCUCGAACAGCGAAAGCGAGAUAUGGCCAAAUCACUG ......((((((....((((((.(((.(.....).)))))))))....((((((...))))))...((....))..............)))))) ( -29.00) >DroGri_CAF1 10259 94 - 1 UAGCGGCAGUGAGAAACUGCAAAUGCUCAAUCAGCGCACCGAGCAGAAUCGUGCGCUGCUCGAGCAGCGUAAACGGGACAUGGCCAAAUCACUA ..((.(((((.....)))))...(((((...((((((((.((......))))))))))...))))))).......((......))......... ( -33.80) >DroMoj_CAF1 6406 92 - 1 --CGGGCAGCGAGAAGUUGCAAAUGCUGAACCAGCGCACCGAGCAAAAUCGUGCGCUCCUCGAGCAGCGCAAACGCGACAUGGCCAAAUCGCUG --....((((((....((((...((((((...(((((((.((......)))))))))..)).))))..))))..((......))....)))))) ( -31.20) >DroAna_CAF1 8637 94 - 1 GAGCGGCAGCGAGAAACUACAGAUGCUCACCCAGCGUACGGAGCAGAAUCGCGCUCUGCUGGAGCAGCGGAAGCGGGAUCUGGCCAAAUCCCUG ....(((((.......)).(((((.(((..(((((....(((((........))))))))))....((....)))))))))))))......... ( -31.70) >consensus CAGCGGCAGCGAGAAACUGCAAAUGCUCAAACAGCGCACCGAACAGAAUCGUGCGCUGCUCGAGCAGCGCAAACGGGACAUGGCCAAAUCACUG ....(((..........(((...(((((...((((((((.((......))))))))))...)))))..)))...........)))......... (-21.84 = -22.73 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:38 2006