
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,808,265 – 3,808,376 |
| Length | 111 |
| Max. P | 0.743144 |

| Location | 3,808,265 – 3,808,376 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -15.82 |
| Energy contribution | -15.38 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
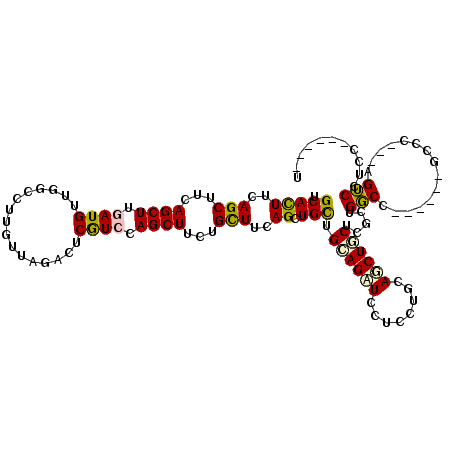
>X_DroMel_CAF1 3808265 111 + 22224390 UGCAUUUCAGCUUUAGCUUUAUGUUGGCCUUGUUAGUUUCGUCCAGCUUUAGCUUCAGCUGCUGCAGAUCCUCCUGCAUCUGCUUGCUGGCG------GCAC---UGUCCUCCGAAUGGU ..((((((((((..((((....(((((.(...........).)))))...))))..)))))..((((..((.((.(((......))).)).)------)..)---))).....))))).. ( -31.00) >DroPse_CAF1 6816 108 + 1 UGCACUUUAGCUUCAGCUUGAUGUUGACCUUGUUCGACUCGUCCAGCUUCUGCUUCAGCUGUUGAAGUUCUUCCUUCAGCUGCUCGCUGGCC------GCCGAGGAGUCCUCC------U .((......)).(((((.....)))))........(((((.((..(((...((...(((.(((((((......))))))).))).)).))).------...)).)))))....------. ( -30.30) >DroGri_CAF1 9382 107 + 1 UGCAUUUGAGCUUCAGCUUGAUGUUGCUCUUGUUGCACUCAUCCAGCUUCUGUUUGAGCUGUUGCAGAUCCGUUUGUAGCUGCUCACUGCCAAUGGAAUCGC---UGGCA---------- ((((...((((..((......))..))))....)))).....(((((((((((((((((.((((((((....)))))))).))))).....)))))))..))---)))..---------- ( -36.40) >DroYak_CAF1 7188 111 + 1 UGCAUUUCAGCUUUAGCUUUAUGUUGGCCUUGUUAGUUUCGUCCAGCUUUUGCUUCAGUUGCUGCAGAUCCUCAUGCAUCUGCUUGCUGGCA------GCAC---UGUCCUCCAAAUGGU .(((...(((((..(((.....(((((.(...........).)))))....)))..)))))..((((((.(....).)))))).))).((((------....---))))..((....)). ( -25.50) >DroAna_CAF1 7518 111 + 1 UGCACUUGAGCUUCAGCUUGAUGUUGGCCUUGUUGGACUCGUCCAGCUUCUGCUUAAGCUGCUGGAGCUCCUCCUGCAACUCCUUGCUGGCC------ACCG---AGUCCUCCAGGGCGU .(((.(..(((....)))..))))..((((((..(((((((.(((((....((....))(((.((((...)))).))).......)))))..------..))---)))))..)))))).. ( -46.60) >DroPer_CAF1 6828 108 + 1 UGCACUUUAGCUUCAGCUUGAUGUUGACCUUGUUCGACUCGUCCAGCUUCUGCUUCAGCUGUUGAAGUUCUUCCUUCAGCUGCUCGCUGGCC------GCCGAGGAGUCCUCC------U .((......)).(((((.....)))))........(((((.((..(((...((...(((.(((((((......))))))).))).)).))).------...)).)))))....------. ( -30.30) >consensus UGCACUUCAGCUUCAGCUUGAUGUUGGCCUUGUUAGACUCGUCCAGCUUCUGCUUCAGCUGCUGCAGAUCCUCCUGCAGCUGCUCGCUGGCC______GCCC___AGUCCUCC______U .(((((..(((...((((.((((................)))).))))...)))..)).))).((((((........)))))).....(((...............)))........... (-15.82 = -15.38 + -0.44)

| Location | 3,808,265 – 3,808,376 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -14.37 |
| Energy contribution | -15.07 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.743144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
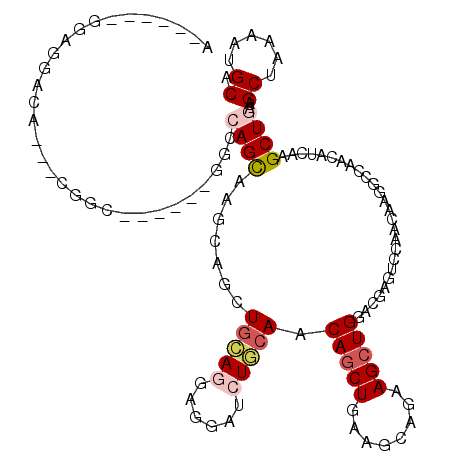
>X_DroMel_CAF1 3808265 111 - 22224390 ACCAUUCGGAGGACA---GUGC------CGCCAGCAAGCAGAUGCAGGAGGAUCUGCAGCAGCUGAAGCUAAAGCUGGACGAAACUAACAAGGCCAACAUAAAGCUAAAGCUGAAAUGCA ....(((((..(..(---((..------(((((((..((((((.(....).))))))...(((....)))...))))).))..)))..)....)).......(((....))))))..... ( -31.20) >DroPse_CAF1 6816 108 - 1 A------GGAGGACUCCUCGGC------GGCCAGCGAGCAGCUGAAGGAAGAACUUCAACAGCUGAAGCAGAAGCUGGACGAGUCGAACAAGGUCAACAUCAAGCUGAAGCUAAAGUGCA .------.((.((((..(((((------..(((((...((((((...(((....)))..))))))........)))))....)))))....))))....)).(((....)))........ ( -32.20) >DroGri_CAF1 9382 107 - 1 ----------UGCCA---GCGAUUCCAUUGGCAGUGAGCAGCUACAAACGGAUCUGCAACAGCUCAAACAGAAGCUGGAUGAGUGCAACAAGAGCAACAUCAAGCUGAAGCUCAAAUGCA ----------(((((---(.(....).)))))).(((((((((................(((((........)))))((((..(((.......))).)))).))))...)))))...... ( -29.00) >DroYak_CAF1 7188 111 - 1 ACCAUUUGGAGGACA---GUGC------UGCCAGCAAGCAGAUGCAUGAGGAUCUGCAGCAACUGAAGCAAAAGCUGGACGAAACUAACAAGGCCAACAUAAAGCUAAAGCUGAAAUGCA .....((((..(..(---((..------(((((((..((((((.(....).)))))).((.......))....))))).))..)))..)....))))(((..(((....)))...))).. ( -29.40) >DroAna_CAF1 7518 111 - 1 ACGCCCUGGAGGACU---CGGU------GGCCAGCAAGGAGUUGCAGGAGGAGCUCCAGCAGCUUAAGCAGAAGCUGGACGAGUCCAACAAGGCCAACAUCAAGCUGAAGCUCAAGUGCA ..(((.((..(((((---((..------..(((((..((((((.(....).))))))....((....))....))))).)))))))..)).)))...(((..(((....)))...))).. ( -44.40) >DroPer_CAF1 6828 108 - 1 A------GGAGGACUCCUCGGC------GGCCAGCGAGCAGCUGAAGGAAGAACUUCAACAGCUGAAGCAGAAGCUGGACGAGUCGAACAAGGUCAACAUCAAGCUGAAGCUAAAGUGCA .------.((.((((..(((((------..(((((...((((((...(((....)))..))))))........)))))....)))))....))))....)).(((....)))........ ( -32.20) >consensus A______GGAGGACA___CGGC______GGCCAGCAAGCAGCUGCAGGAGGAUCUGCAACAGCUGAAGCAGAAGCUGGACGAGUCCAACAAGGCCAACAUCAAGCUGAAGCUAAAAUGCA ...............................((((.......(((((......))))).(((((........)))))..........................))))..((......)). (-14.37 = -15.07 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:34 2006