
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,744,614 – 3,744,788 |
| Length | 174 |
| Max. P | 0.978880 |

| Location | 3,744,614 – 3,744,733 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -33.00 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3744614 119 + 22224390 UUCCGCCGCCCACAG-AUUUCUGUGGUCAUGCAAUCCUGGCAGCCAUUAGCGGAAUAUGGCUAAGAUGACGCCCACACGCAAAAGCACAUAGCAAUUUGCGUGUGAUGUGCAGGAAAGCA .......(.((((((-....)))))).).(((..(((((..((((((.........)))))).......(((.((((((((((.((.....))..))))))))))..))))))))..))) ( -43.90) >DroSec_CAF1 17514 116 + 1 UUCCGCCGCACACAG-AUUUCUGUGGUCAUGCAAUCCUGGCAGCCAUUAGCGAAAUAUGGCUAAGAUG-CGCCCACACGCAAAAGCACACAGCAAUUUGCGUGUGC--UGCAGGAAAGCA ((((...((.(((((-....)))))))..((((.....(((.((..(((((........)))))...)-))))((((((((((.((.....))..)))))))))).--)))))))).... ( -41.50) >DroEre_CAF1 18187 118 + 1 UUCCGCCGCCCACAAAAUUUUUGUGGUCAUGCAAUCCUGGCUGCCAUUAGCGAAAUAUGGCUAAGAUGACGCCCACACGCAAAAGCACACAGCAAUUUGCGUGUCC--UGCAGGAAAGCA ((((...(.((((((.....)))))).).((((.....(((..(..(((((........)))))...)..))).(((((((((.((.....))..)))))))))..--)))))))).... ( -36.10) >DroYak_CAF1 18080 117 + 1 AUCCGCCGCCCACAA-AUUUUUGUGGUCAUGCAAUGCUGGCAGCCAUUAGCGAAAUAUGGCUAAGAUGACACCCACACGCAAAUGCACACAGAAAUUUGCGUGUGC--AGCAGGAAAUCA .(((((.(.((((((-....)))))).)..))..(((((..((((((.........))))))...........(((((((((((..........))))))))))))--)))))))..... ( -40.80) >consensus UUCCGCCGCCCACAA_AUUUCUGUGGUCAUGCAAUCCUGGCAGCCAUUAGCGAAAUAUGGCUAAGAUGACGCCCACACGCAAAAGCACACAGCAAUUUGCGUGUGC__UGCAGGAAAGCA .(((...(.((((((.....)))))).).(((......(((.....(((((........)))))......)))((((((((((.((.....))..))))))))))....))))))..... (-33.00 = -33.50 + 0.50)

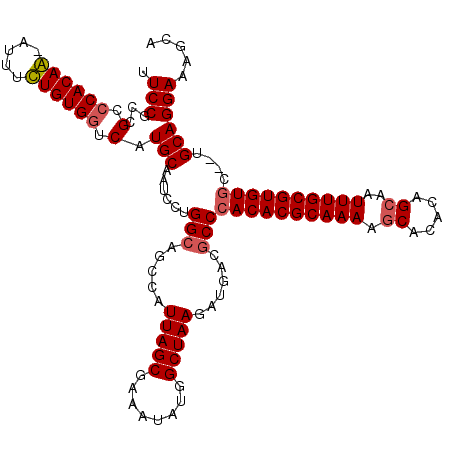
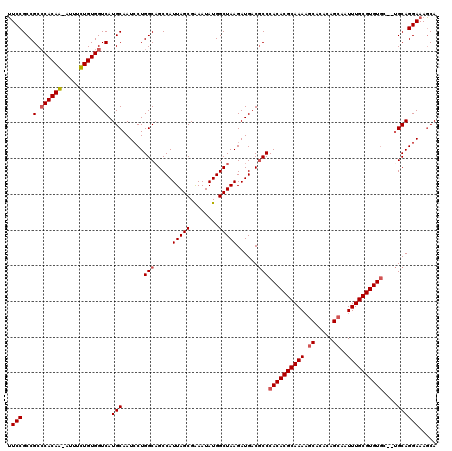
| Location | 3,744,614 – 3,744,733 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -38.24 |
| Energy contribution | -38.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3744614 119 - 22224390 UGCUUUCCUGCACAUCACACGCAAAUUGCUAUGUGCUUUUGCGUGUGGGCGUCAUCUUAGCCAUAUUCCGCUAAUGGCUGCCAGGAUUGCAUGACCACAGAAAU-CUGUGGGCGGCGGAA (((..((((......((((((((((..((.....)).))))))))))((((((((..((((........))))))))).)))))))..)))...((((((....-))))))......... ( -49.30) >DroSec_CAF1 17514 116 - 1 UGCUUUCCUGCA--GCACACGCAAAUUGCUGUGUGCUUUUGCGUGUGGGCG-CAUCUUAGCCAUAUUUCGCUAAUGGCUGCCAGGAUUGCAUGACCACAGAAAU-CUGUGUGCGGCGGAA (((..(((((((--(((((((((((..((.....)).))))))))).(((.-.......)))..............)))).)))))..)))((..(((((....-)))))..))...... ( -46.30) >DroEre_CAF1 18187 118 - 1 UGCUUUCCUGCA--GGACACGCAAAUUGCUGUGUGCUUUUGCGUGUGGGCGUCAUCUUAGCCAUAUUUCGCUAAUGGCAGCCAGGAUUGCAUGACCACAAAAAUUUUGUGGGCGGCGGAA (((..((((...--..(((((((((..((.....)).))))))))).((((((((..((((........))))))))).)))))))..)))...((((((.....))))))......... ( -43.40) >DroYak_CAF1 18080 117 - 1 UGAUUUCCUGCU--GCACACGCAAAUUUCUGUGUGCAUUUGCGUGUGGGUGUCAUCUUAGCCAUAUUUCGCUAAUGGCUGCCAGCAUUGCAUGACCACAAAAAU-UUGUGGGCGGCGGAU .......(((((--((...(((((((((.((((..((..(((((((.((((((((..((((........))))))))).))).))).))))))..)))).))))-))))).))))))).. ( -45.40) >consensus UGCUUUCCUGCA__GCACACGCAAAUUGCUGUGUGCUUUUGCGUGUGGGCGUCAUCUUAGCCAUAUUUCGCUAAUGGCUGCCAGGAUUGCAUGACCACAAAAAU_CUGUGGGCGGCGGAA .....((((......((((((((((..((.....)).))))))))))((((((((..((((........))))))))).)))))))((((....((((((.....))))))...)))).. (-38.24 = -38.80 + 0.56)

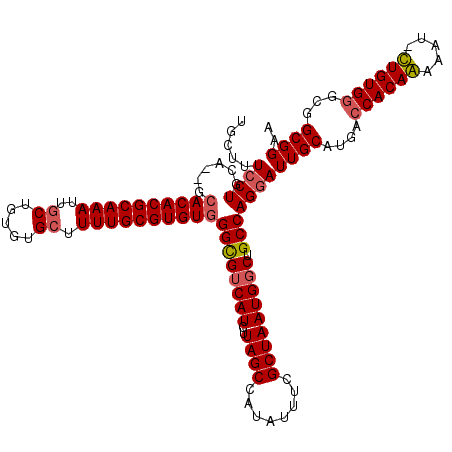
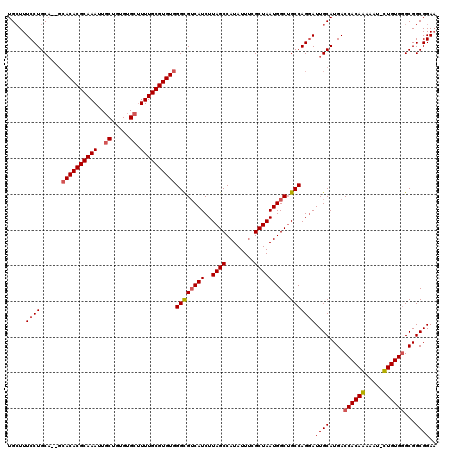
| Location | 3,744,653 – 3,744,771 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.09 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -32.71 |
| Energy contribution | -32.77 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3744653 118 - 22224390 --ACUGCUUGAGGGCUGCGUGUGUGGCUUCGAUACUCGAAUGCUUUCCUGCACAUCACACGCAAAUUGCUAUGUGCUUUUGCGUGUGGGCGUCAUCUUAGCCAUAUUCCGCUAAUGGCUG --..........(((((((.((((((((..(((((.(...(((......)))..(((((((((((..((.....)).)))))))))))).)).)))..))))))))..)))....)))). ( -41.80) >DroSec_CAF1 17553 115 - 1 --ACUGCUUGAGGGCUGCGUGUGUGGUUUCGAUACUCGAAUGCUUUCCUGCA--GCACACGCAAAUUGCUGUGUGCUUUUGCGUGUGGGCG-CAUCUUAGCCAUAUUUCGCUAAUGGCUG --.((((..((((((..((.((((.(...).)))).))...))))))..)))--)((((((((((..((.....)).))))))))))(((.-(((..((((........)))))))))). ( -43.80) >DroEre_CAF1 18227 116 - 1 --ACUGCUUGAGGGCUGCGUGUGUGGUUUCGUUACUCGAAUGCUUUCCUGCA--GGACACGCAAAUUGCUGUGUGCUUUUGCGUGUGGGCGUCAUCUUAGCCAUAUUUCGCUAAUGGCAG --.((((..((((((..((.(((.........))).))...))))))..)))--).(((((((((..((.....)).)))))))))....(((((..((((........))))))))).. ( -40.10) >DroYak_CAF1 18119 118 - 1 ACCCUGCUUGAGGGCUGCGUGUGUGGCUUCGUUACUCGAAUGAUUUCCUGCU--GCACACGCAAAUUUCUGUGUGCAUUUGCGUGUGGGUGUCAUCUUAGCCAUAUUUCGCUAAUGGCUG ((((.....((((..((((((((..((.(((((.....)))))......)).--.))))))))..)))).(..(((....)))..))))).......(((((((.........))))))) ( -40.60) >consensus __ACUGCUUGAGGGCUGCGUGUGUGGCUUCGAUACUCGAAUGCUUUCCUGCA__GCACACGCAAAUUGCUGUGUGCUUUUGCGUGUGGGCGUCAUCUUAGCCAUAUUUCGCUAAUGGCUG .....(((....(((.....(((((((((((.....)))).........((....((((((((((..((.....)).)))))))))).)).........)))))))...)))...))).. (-32.71 = -32.77 + 0.06)


| Location | 3,744,693 – 3,744,788 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -20.61 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3744693 95 + 22224390 AAAAGCACAUAGCAAUUUGCGUGUGAUGUGCAGGAAAGCAUUCGAGUAUCGAAGCCACACACGCAGCCCUCAAGCAGU------------------GCAAGCAGUGGAAGCAG------- ....((((.........((((((((...(((......)))((((.....))))....))))))))((......)).))------------------))..((.......))..------- ( -27.40) >DroSec_CAF1 17592 93 + 1 AAAAGCACACAGCAAUUUGCGUGUGC--UGCAGGAAAGCAUUCGAGUAUCGAAACCACACACGCAGCCCUCAAGCAGU------------------GCAAGCAGUGGAAGCAG------- ....((.(((.((....((((((((.--(((......)))((((.....))))....))))))))((.((.....)).------------------))..)).)))...))..------- ( -26.90) >DroEre_CAF1 18267 100 + 1 AAAAGCACACAGCAAUUUGCGUGUCC--UGCAGGAAAGCAUUCGAGUAACGAAACCACACACGCAGCCCUCAAGCAGU------------------GCAAGCAGUGAAAGCAGUGUACAU ....((((.........(((((((..--(((......)))((((.....)))).....)))))))((......)).))------------------))..(((.((....)).))).... ( -24.90) >DroYak_CAF1 18159 118 + 1 AAAUGCACACAGAAAUUUGCGUGUGC--AGCAGGAAAUCAUUCGAGUAACGAAGCCACACACGCAGCCCUCAAGCAGGGUCAACAUGGCAAGUAUGGCAAGCAGUGCAAGCAGUGGACAU ..(((..(((.......((((((((.--.((..(....).((((.....))))))..))))))))(((((.....))))).......((..((((.(....).))))..)).)))..))) ( -31.70) >consensus AAAAGCACACAGCAAUUUGCGUGUGC__UGCAGGAAAGCAUUCGAGUAACGAAACCACACACGCAGCCCUCAAGCAGU__________________GCAAGCAGUGGAAGCAG_______ ....((.(((.((....((((((((....((......)).((((.....))))....))))))))((......)).........................)).)))...))......... (-20.61 = -21.18 + 0.56)


| Location | 3,744,693 – 3,744,788 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.70 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -20.85 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3744693 95 - 22224390 -------CUGCUUCCACUGCUUGC------------------ACUGCUUGAGGGCUGCGUGUGUGGCUUCGAUACUCGAAUGCUUUCCUGCACAUCACACGCAAAUUGCUAUGUGCUUUU -------..(((..((..((....------------------...)).))..)))((((((((((..((((.....))))(((......))))).))))))))....((.....)).... ( -27.60) >DroSec_CAF1 17592 93 - 1 -------CUGCUUCCACUGCUUGC------------------ACUGCUUGAGGGCUGCGUGUGUGGUUUCGAUACUCGAAUGCUUUCCUGCA--GCACACGCAAAUUGCUGUGUGCUUUU -------.(((...........))------------------).(((..((((((..((.((((.(...).)))).))...))))))..)))--((((((((.....)).)))))).... ( -29.10) >DroEre_CAF1 18267 100 - 1 AUGUACACUGCUUUCACUGCUUGC------------------ACUGCUUGAGGGCUGCGUGUGUGGUUUCGUUACUCGAAUGCUUUCCUGCA--GGACACGCAAAUUGCUGUGUGCUUUU ..((((((.((......(((.((.------------------.((((..((((((..((.(((.........))).))...))))))..)))--)..)).)))....)).)))))).... ( -34.20) >DroYak_CAF1 18159 118 - 1 AUGUCCACUGCUUGCACUGCUUGCCAUACUUGCCAUGUUGACCCUGCUUGAGGGCUGCGUGUGUGGCUUCGUUACUCGAAUGAUUUCCUGCU--GCACACGCAAAUUUCUGUGUGCAUUU (((..(((.((..((...))..)).................((((.....)))).((((((((..((.(((((.....)))))......)).--.)))))))).......)))..))).. ( -34.00) >consensus _______CUGCUUCCACUGCUUGC__________________ACUGCUUGAGGGCUGCGUGUGUGGCUUCGAUACUCGAAUGCUUUCCUGCA__GCACACGCAAAUUGCUGUGUGCUUUU .........((...(((.((.........................(((....)))(((((((((.((((((.....)))).........))...)))))))))....)).))).)).... (-20.85 = -21.60 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:05:06 2006