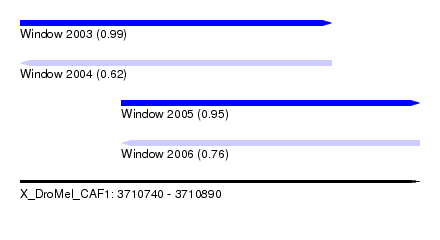
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,710,740 – 3,710,890 |
| Length | 150 |
| Max. P | 0.989435 |

| Location | 3,710,740 – 3,710,857 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.36 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
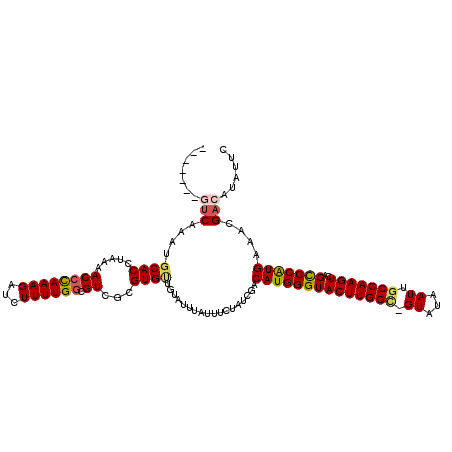
>X_DroMel_CAF1 3710740 117 + 22224390 AUCUCCUCUACAUUUCUUUAUCAGUGUCCCACCAGUGAGAAUAUGUCGUUUCAUGGGCUGACUUGGCAAUUAUAC-GCCAAGUACCCAUGUCGAUAGAAAUAAGUACAGCACACAACG .....((.((((((((....(((.((......)).))).....(((((...((((((...(((((((........-))))))).)))))).))))))))))..))).))......... ( -32.60) >DroSec_CAF1 2334 117 + 1 AUCUCCUCUACAUUUCUUUAUCAAUGUCCCACCAGUGAGAAUAUGUCGUUUCAUGGACUGACUUGGCAAUUAUAC-GCCAAGUACCCAUGUCGAUAGAAUUAAAUACAACACGCGACG .((((((..(((((........)))))......)).))))....(((((..(((((....(((((((........-)))))))..)))))((....))..............))))). ( -26.80) >DroSim_CAF1 2334 117 + 1 AUCUCCUCUACAUUUCUUUAUCAAUGUCCCACCAGUGAGAAUAUGUCGUUUCAUGGACUGACUUGGCAAUUAUAC-GCCAAGUACCCAUGUCGAUAGAAUUAAAUACAACACGCGACG .((((((..(((((........)))))......)).))))....(((((..(((((....(((((((........-)))))))..)))))((....))..............))))). ( -26.80) >DroEre_CAF1 2355 118 + 1 AUCUCGUCUACGUUUAUUUAUCGAUGUCCCACCAUUGAGAAUAUGUCGUUUCACGGGCUGACUUGGCAAUUAUACCACCAAGUACCCAUGUCGAUAGAAAUUUAAAAACCACCCGACG ....((((......(((((.((((((......)))))))))))(((((...((.(((...((((((...........)))))).))).)).)))))..................)))) ( -26.50) >DroYak_CAF1 2352 118 + 1 AUCUCGUCUAUGUUUCUUUAUCGAUGUUCCACCAUUGAGAAUAUGUCGUUUCAUGGGCUGACUUGGCAAUUAUACCCCCAAGUACCCAUGUCGAUAGAAAUUUAUACAACACCCGACG ....((((..((((......((((((......))))))((((.(((((...((((((...((((((...........)))))).)))))).)))))...))))....))))...)))) ( -30.10) >consensus AUCUCCUCUACAUUUCUUUAUCAAUGUCCCACCAGUGAGAAUAUGUCGUUUCAUGGGCUGACUUGGCAAUUAUAC_GCCAAGUACCCAUGUCGAUAGAAAUAAAUACAACACCCGACG .((((((..(((((........)))))......)).))))...(((((...((((((...(((((((.........))))))).)))))).)))))...................... (-23.64 = -23.36 + -0.28)

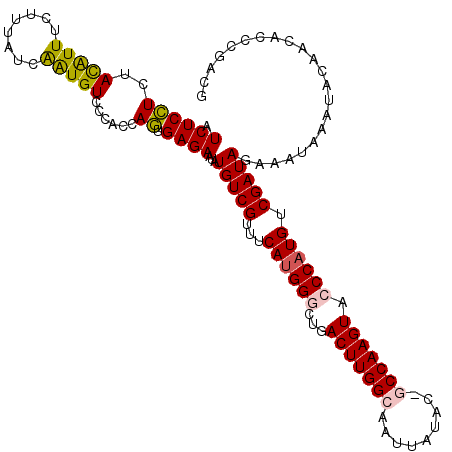
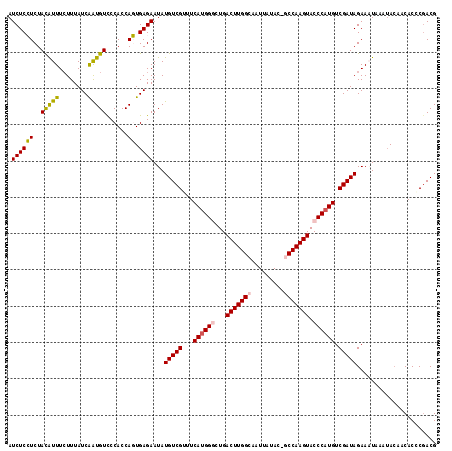
| Location | 3,710,740 – 3,710,857 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.48 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -24.42 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3710740 117 - 22224390 CGUUGUGUGCUGUACUUAUUUCUAUCGACAUGGGUACUUGGC-GUAUAAUUGCCAAGUCAGCCCAUGAAACGACAUAUUCUCACUGGUGGGACACUGAUAAAGAAAUGUAGAGGAGAU ....((.(.((.(((..((((((((((.((((((((((((((-(......))))))))..)))))))...........(((((....)))))...))))..))))))))).)).).)) ( -37.00) >DroSec_CAF1 2334 117 - 1 CGUCGCGUGUUGUAUUUAAUUCUAUCGACAUGGGUACUUGGC-GUAUAAUUGCCAAGUCAGUCCAUGAAACGACAUAUUCUCACUGGUGGGACAUUGAUAAAGAAAUGUAGAGGAGAU .(((..(((((((.......((....))((((((.(((((((-(......))))))))...))))))..))))))).(((((.((....))(((((........))))).)))))))) ( -31.30) >DroSim_CAF1 2334 117 - 1 CGUCGCGUGUUGUAUUUAAUUCUAUCGACAUGGGUACUUGGC-GUAUAAUUGCCAAGUCAGUCCAUGAAACGACAUAUUCUCACUGGUGGGACAUUGAUAAAGAAAUGUAGAGGAGAU .(((..(((((((.......((....))((((((.(((((((-(......))))))))...))))))..))))))).(((((.((....))(((((........))))).)))))))) ( -31.30) >DroEre_CAF1 2355 118 - 1 CGUCGGGUGGUUUUUAAAUUUCUAUCGACAUGGGUACUUGGUGGUAUAAUUGCCAAGUCAGCCCGUGAAACGACAUAUUCUCAAUGGUGGGACAUCGAUAAAUAAACGUAGACGAGAU .(((((((((...........)))))..(((((((((((((..((...))..))))))..)))))))...))))....((((..(((((...)))))..........(....))))). ( -34.50) >DroYak_CAF1 2352 118 - 1 CGUCGGGUGUUGUAUAAAUUUCUAUCGACAUGGGUACUUGGGGGUAUAAUUGCCAAGUCAGCCCAUGAAACGACAUAUUCUCAAUGGUGGAACAUCGAUAAAGAAACAUAGACGAGAU ((((..(((((.(((..((((((((((.(((((((((((((.(((...))).))))))..)))))))....((.......))..)))))))).))..)))....))))).)))).... ( -35.20) >consensus CGUCGCGUGUUGUAUUUAUUUCUAUCGACAUGGGUACUUGGC_GUAUAAUUGCCAAGUCAGCCCAUGAAACGACAUAUUCUCACUGGUGGGACAUUGAUAAAGAAAUGUAGAGGAGAU .((((.(((((((.......((....))((((((((((((((.((...)).)))))))..)))))))..)))))))..(((((....)))))...))))................... (-24.42 = -24.50 + 0.08)

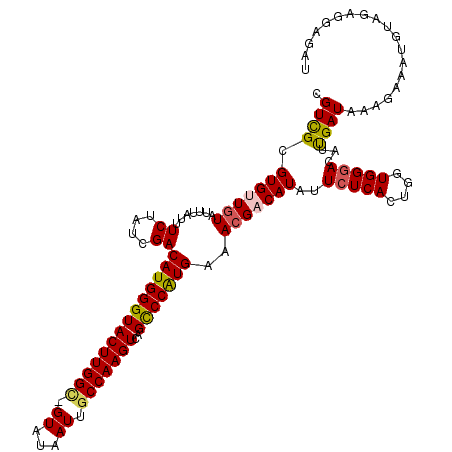

| Location | 3,710,778 – 3,710,890 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -25.38 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3710778 112 + 22224390 GAAUAUGUCGUUUCAUGGGCUGACUUGGCAAUUAUAC-GCCAAGUACCCAUGUCGAUAGAAAUAAGUACAGCACACAACGCCAAAAGAUCUUUGGGUUUUAGGUGCAUUUGAC------- .....(((((...((((((...(((((((........-))))))).)))))).)))))............((((..(((.(((((.....))))))))....)))).......------- ( -37.70) >DroSec_CAF1 2372 112 + 1 GAAUAUGUCGUUUCAUGGACUGACUUGGCAAUUAUAC-GCCAAGUACCCAUGUCGAUAGAAUUAAAUACAACACGCGACGCCAAAAGAUCUUUGGGUUUUAGGUGCAUUUGAC------- .....(((((...(((((....(((((((........-)))))))..))))).)))))...((((((.((...(..(((.(((((.....))))))))...).)).)))))).------- ( -30.90) >DroSim_CAF1 2372 112 + 1 GAAUAUGUCGUUUCAUGGACUGACUUGGCAAUUAUAC-GCCAAGUACCCAUGUCGAUAGAAUUAAAUACAACACGCGACGCCAAAAGAUCUUUGGGUUUUAGGUGCAUAUGAC------- .....(((((...(((((....(((((((........-)))))))..))))).)))))..................(.((((..((((((....)))))).))))).......------- ( -29.90) >DroEre_CAF1 2393 120 + 1 GAAUAUGUCGUUUCACGGGCUGACUUGGCAAUUAUACCACCAAGUACCCAUGUCGAUAGAAAUUUAAAAACCACCCGACGCCAAAAGAUCUUUGGGUUUUAGGUGCAUUUGAACGUACAA .....(((((...((.(((...((((((...........)))))).))).)).)))))....((((((...((((.(((.(((((.....))))))))...))))..))))))....... ( -28.50) >DroYak_CAF1 2390 116 + 1 GAAUAUGUCGUUUCAUGGGCUGACUUGGCAAUUAUACCCCCAAGUACCCAUGUCGAUAGAAAUUUAUACAACACCCGACGUCAAAAGAUCUUUAGGUUUUAGGUGCUUUUGGACGU---- .....(((((...((((((...((((((...........)))))).)))))).)))))...................((((((((((((((.........)))).))))).)))))---- ( -31.00) >consensus GAAUAUGUCGUUUCAUGGGCUGACUUGGCAAUUAUAC_GCCAAGUACCCAUGUCGAUAGAAAUAAAUACAACACCCGACGCCAAAAGAUCUUUGGGUUUUAGGUGCAUUUGAC_______ .....(((((...((((((...(((((((.........))))))).)))))).)))))....................((((..((((((....)))))).))))............... (-25.38 = -26.22 + 0.84)



| Location | 3,710,778 – 3,710,890 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -22.22 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3710778 112 - 22224390 -------GUCAAAUGCACCUAAAACCCAAAGAUCUUUUGGCGUUGUGUGCUGUACUUAUUUCUAUCGACAUGGGUACUUGGC-GUAUAAUUGCCAAGUCAGCCCAUGAAACGACAUAUUC -------(((....((((....(((((((((...)))))).)))..))))..................((((((((((((((-(......))))))))..)))))))....)))...... ( -37.50) >DroSec_CAF1 2372 112 - 1 -------GUCAAAUGCACCUAAAACCCAAAGAUCUUUUGGCGUCGCGUGUUGUAUUUAAUUCUAUCGACAUGGGUACUUGGC-GUAUAAUUGCCAAGUCAGUCCAUGAAACGACAUAUUC -------((((((((((.(....((((((((...)))))).)).....).)))))))...........((((((.(((((((-(......))))))))...))))))....)))...... ( -30.70) >DroSim_CAF1 2372 112 - 1 -------GUCAUAUGCACCUAAAACCCAAAGAUCUUUUGGCGUCGCGUGUUGUAUUUAAUUCUAUCGACAUGGGUACUUGGC-GUAUAAUUGCCAAGUCAGUCCAUGAAACGACAUAUUC -------(((((((((((((((((..........)))))).)).))))))..................((((((.(((((((-(......))))))))...))))))....)))...... ( -30.40) >DroEre_CAF1 2393 120 - 1 UUGUACGUUCAAAUGCACCUAAAACCCAAAGAUCUUUUGGCGUCGGGUGGUUUUUAAAUUUCUAUCGACAUGGGUACUUGGUGGUAUAAUUGCCAAGUCAGCCCGUGAAACGACAUAUUC .(((.((((......(((((...((((((((...)))))).)).)))))...................(((((((((((((..((...))..))))))..))))))).)))))))..... ( -35.90) >DroYak_CAF1 2390 116 - 1 ----ACGUCCAAAAGCACCUAAAACCUAAAGAUCUUUUGACGUCGGGUGUUGUAUAAAUUUCUAUCGACAUGGGUACUUGGGGGUAUAAUUGCCAAGUCAGCCCAUGAAACGACAUAUUC ----(((((.(((((...((.........))..))))))))))...(((((((.......((....))(((((((((((((.(((...))).))))))..)))))))..))))))).... ( -30.80) >consensus _______GUCAAAUGCACCUAAAACCCAAAGAUCUUUUGGCGUCGCGUGUUGUAUUUAUUUCUAUCGACAUGGGUACUUGGC_GUAUAAUUGCCAAGUCAGCCCAUGAAACGACAUAUUC .......(((....((((.....((((((((...)))))).))...))))..................((((((((((((((.((...)).)))))))..)))))))....)))...... (-22.22 = -22.62 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:56 2006