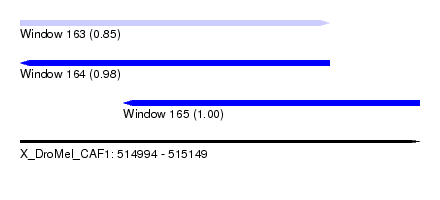
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 514,994 – 515,149 |
| Length | 155 |
| Max. P | 0.995269 |

| Location | 514,994 – 515,114 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -24.90 |
| Energy contribution | -25.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 514994 120 + 22224390 AGUGUGCAAUUGAAAUGCGCGCCAACGCAUGUUCUCAUAUGCGGAGCGUUUAGCAGCCCUUAAAAAGCUUUUCAUGUUGCUUUUAAAAAGCUACGACGGGUGUGGAAAUUAGAUUUGUGA .(((((((.......)))))))...((((.(((..(((((.((..((.....))(((..((.((((((..........)))))).))..)))....)).)))))..)))......)))). ( -30.50) >DroSec_CAF1 1038 117 + 1 ---GUACAAUUGAAAUGCGCGCCAACGCAUUUUCUCAUAUGCGGUGCGCUUCGCAGCACUUAAAAAGCUUUUCAUUUUGCUUUUUAAAAGCUACGACGGGUGUGAAAAUCAGAUUUGUGA ---.(((((((((....((((((..(((((........)))))...((..(((.(((..(((((((((..........)))))))))..))).)))))))))))....))))..))))). ( -35.00) >DroSim_CAF1 1052 117 + 1 ---GUACAAUUGAAAUGCGCGCCAACGCAUUUUCUCAUAUGCGGUGCGUUUCGCAGCACUUAAAAAGCUUUUCAUUUUGCUUUUAAAAAGCUACGACGGGUGUGAAAAUCAGAUUUGUGA ---.(((((((((....((((((..(((((........)))))...((..(((.(((..((.((((((..........)))))).))..))).)))))))))))....))))..))))). ( -31.10) >DroEre_CAF1 887 100 + 1 ----------------GCGCGCCAACGCAUUGU-UAAUAUG---UGCGUGUUGCAGCACUUGAAAAGCUUUUCAUGUUGCUUUUCAAAAGCUACGACGGGUGUGGGAAUCCGAUUUUUGA ----------------.((((((.((((((...-......)---)))))((((.(((..(((((((((..........)))))))))..))).)))).))))))................ ( -34.30) >consensus ___GUACAAUUGAAAUGCGCGCCAACGCAUUUUCUCAUAUGCGGUGCGUUUCGCAGCACUUAAAAAGCUUUUCAUGUUGCUUUUAAAAAGCUACGACGGGUGUGAAAAUCAGAUUUGUGA .................((((((..(((((........)))))...((..(((.(((..(((((((((..........)))))))))..))).)))))))))))................ (-24.90 = -25.90 + 1.00)

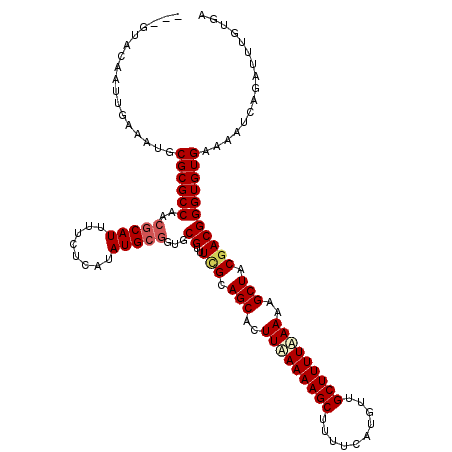
| Location | 514,994 – 515,114 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -23.62 |
| Energy contribution | -24.05 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 514994 120 - 22224390 UCACAAAUCUAAUUUCCACACCCGUCGUAGCUUUUUAAAAGCAACAUGAAAAGCUUUUUAAGGGCUGCUAAACGCUCCGCAUAUGAGAACAUGCGUUGGCGCGCAUUUCAAUUGCACACU ......................(((.(((((((((.((((((..........)))))).)))))))))...)))....(((..(((((...(((((....))))))))))..)))..... ( -28.30) >DroSec_CAF1 1038 117 - 1 UCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUAAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAGCGCACCGCAUAUGAGAAAAUGCGUUGGCGCGCAUUUCAAUUGUAC--- ..((((...(((..(.(.......(((((((.((((((((((..........)))))))))).))))))).((((..(((((........)))))...))))).)..))).))))..--- ( -35.70) >DroSim_CAF1 1052 117 - 1 UCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUUAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAACGCACCGCAUAUGAGAAAAUGCGUUGGCGCGCAUUUCAAUUGUAC--- ..((((.(((.((...........(((((((.(((.((((((..........)))))).))).)))))))............)).)))((((((((....))))))))...))))..--- ( -28.10) >DroEre_CAF1 887 100 - 1 UCAAAAAUCGGAUUCCCACACCCGUCGUAGCUUUUGAAAAGCAACAUGAAAAGCUUUUCAAGUGCUGCAACACGCA---CAUAUUA-ACAAUGCGUUGGCGCGC---------------- .........((....)).....(((((((((.((((((((((..........)))))))))).)))))...(((((---.......-....))))).))))...---------------- ( -28.00) >consensus UCACAAAUCUGAUUUCCACACCCGUCGUAGCUUUUUAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAACGCACCGCAUAUGAGAAAAUGCGUUGGCGCGCAUUUCAAUUGUAC___ ......................(((((((((.((((((((((..........)))))))))).)))))..(((((.................)))))))))................... (-23.62 = -24.05 + 0.44)

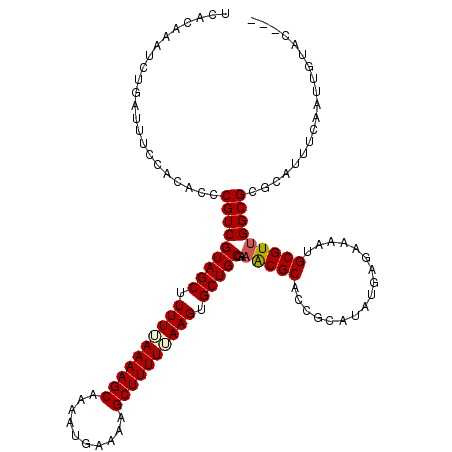
| Location | 515,034 – 515,149 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -19.11 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
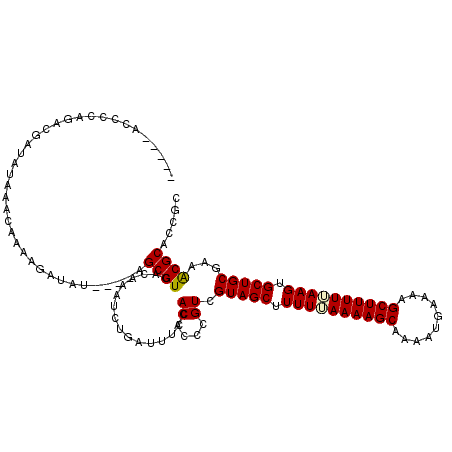
>X_DroMel_CAF1 515034 115 - 22224390 -----ACCCCAGACGAUAUAAAAAAAAGAUAUAUAAAGCGUCACAAAUCUAAUUUCCACACCCGUCGUAGCUUUUUAAAAGCAACAUGAAAAGCUUUUUAAGGGCUGCUAAACGCUCCGC -----......((((.((((...........))))...))))....................(((.(((((((((.((((((..........)))))).)))))))))...)))...... ( -20.40) >DroSec_CAF1 1075 111 - 1 -----ACCCCAGACGAUAUAAACAAAAGAUAU----AGCGUCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUAAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAGCGCACCGC -----......((((.((((.........)))----).))))......................(((((((.((((((((((..........)))))))))).))))))).(((...))) ( -25.80) >DroSim_CAF1 1089 111 - 1 -----ACCCCAGACGAUAUAAACAAAAGAUAU----AGCGUCACAAAUCUGAUUUUCACACCCGUCGUAGCUUUUUAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAACGCACCGC -----......((((.((((.........)))----).))))......................(((((((.(((.((((((..........)))))).))).))))))).......... ( -21.50) >DroEre_CAF1 910 99 - 1 UACAUACACCAA--------------AAAUUA----AGCGUCAAAAAUCGGAUUCCCACACCCGUCGUAGCUUUUGAAAAGCAACAUGAAAAGCUUUUCAAGUGCUGCAACACGCA---C ............--------------......----.((((........((....))......((.(((((.((((((((((..........)))))))))).))))).)))))).---. ( -26.60) >consensus _____ACCCCAGACGAUAUAAACAAAAGAUAU____AGCGUCACAAAUCUGAUUUCCACACCCGUCGUAGCUUUUUAAAAGCAAAAUGAAAAGCUUUUUAAGUGCUGCGAAACGCACCGC .....................................((((................((....)).(((((.((((((((((..........)))))))))).)))))...))))..... (-19.11 = -19.30 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:53 2006