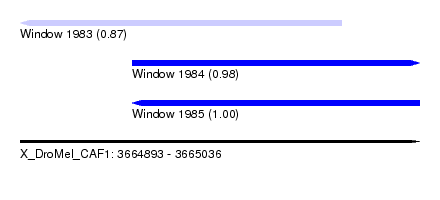
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,664,893 – 3,665,036 |
| Length | 143 |
| Max. P | 0.999449 |

| Location | 3,664,893 – 3,665,008 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -26.45 |
| Energy contribution | -26.45 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3664893 115 - 22224390 UAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCAACACGUCAAACGGCGAGCAGAAUGUUGCUUAUCAAUUUCUU ..((((((((((((.......))))).....((.((((((....)))))).)).............)))))))....((((....))))(((((....)))))............ ( -33.70) >DroSec_CAF1 16712 115 - 1 UAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCAACACGCCAAUUAACGAGCAGAAUGGUGCUUAUCAAUUUCUU ..((((((((((((.......))))).....((.((((((....)))))).)).............)))))))....(((((.((........)).))))).............. ( -31.60) >DroYak_CAF1 19181 115 - 1 AAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCACAUAACGACCACAGAGCAGAAAAAUUCUUAUCAAUUUCUU ..((((((((((((.......))))).....((.((((((....)))))).)).............)))))))...............((..(((.....)))..))........ ( -27.00) >DroAna_CAF1 22546 102 - 1 AAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCUCUUGUCAGCCAC------GAAUUUUUCUUCCC-------C ..((((((((((((.......))))).....((.((((((....)))))).)).............)))))))..............------..............-------. ( -26.00) >consensus AAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCAACACGUCAACCACCGAGCAGAAUGUUGCUUAUCAAUUUCUU ..((((((((((((.......))))).....((.((((((....)))))).)).............))))))).......................................... (-26.45 = -26.45 + -0.00)

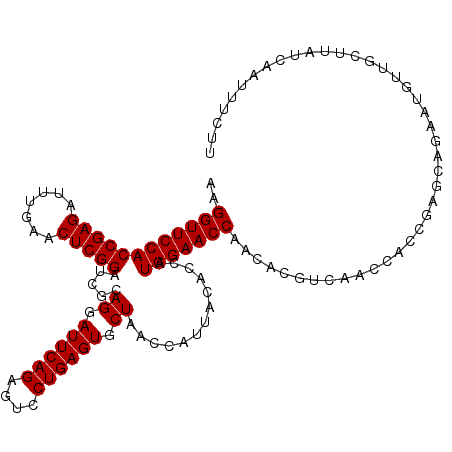

| Location | 3,664,933 – 3,665,036 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -26.74 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3664933 103 + 22224390 UUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUA-ACGAUAUGUUUUCUUUUUUGUUAAAAAA ..(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))((-((((.............))))))..... ( -27.22) >DroSec_CAF1 16752 87 + 1 UUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUA-ACCAUGUGUUUU---------------- ..(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))..-............---------------- ( -25.40) >DroYak_CAF1 19221 99 + 1 UGGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUUGAGGAGAUU-----UUUUUUGUUGAUAAA ..(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))(..(.((((..-----..)))).)..).... ( -27.60) >consensus UUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCCGAGUUCAAAUCUCGGUGGAACCUA_ACGAUAUGUUUU_UUUUUUGUU_A_AAA ..(((((((.(((((........)))))(((((.......)))))....(((((.......))))))))))))............................... (-25.40 = -25.40 + 0.00)

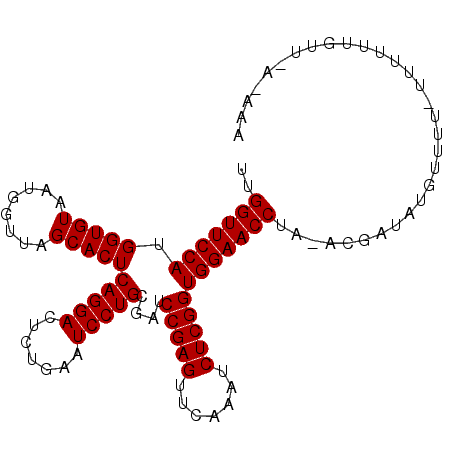

| Location | 3,664,933 – 3,665,036 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -26.60 |
| Energy contribution | -26.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3664933 103 - 22224390 UUUUUUAACAAAAAAGAAAACAUAUCGU-UAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCAA ............................-..((((((((((((.......))))).....((.((((((....)))))).)).............))))))).. ( -26.90) >DroSec_CAF1 16752 87 - 1 ----------------AAAACACAUGGU-UAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCAA ----------------............-..((((((((((((.......))))).....((.((((((....)))))).)).............))))))).. ( -26.90) >DroYak_CAF1 19221 99 - 1 UUUAUCAACAAAAAA-----AAUCUCCUCAAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCCA ...............-----...........((((((((((((.......))))).....((.((((((....)))))).)).............))))))).. ( -26.00) >consensus UUU_U_AACAAAAAA_AAAACAUAUCGU_UAGGUUCCACCGAGAUUUGAACUCGGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCAA ...............................((((((((((((.......))))).....((.((((((....)))))).)).............))))))).. (-26.60 = -26.60 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:35 2006