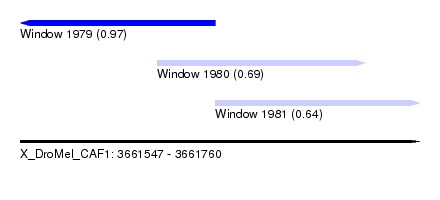
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,661,547 – 3,661,760 |
| Length | 213 |
| Max. P | 0.973844 |

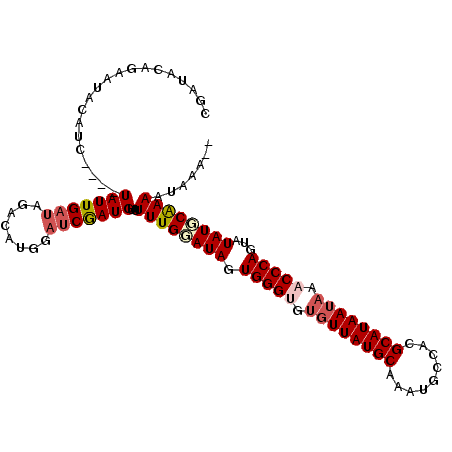
| Location | 3,661,547 – 3,661,651 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.87 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -21.06 |
| Energy contribution | -22.50 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3661547 104 - 22224390 CGAUACAGAAUACAUC----UAUUGAUAGGCAUGGAUCGAUGUUUUUGUAUAGUGGGUGUGUUAUGCAAAUGCCGCGCAUAAUAACCCCAGUAUAUGCAAAAAAAAAA ((((.((......(((----....))).....)).))))...((((((((((.((((..((((((((.........))))))))..))))...))))))))))..... ( -27.90) >DroSec_CAF1 13480 98 - 1 CGAUACAGAAUUCAUC----UAUUGAGAGGCAUGGACCGAUGAUUUUGGAUAGUGGGUGUGUUAUGCAAAUGCCGCGCAUAAUCACCCCAGUAUAUGCAAAA------ ..........((((..----...))))..((((...((((.....))))(((.((((.(((((((((.........)))))).))))))).)))))))....------ ( -26.60) >DroEre_CAF1 14356 102 - 1 CGAUACAGAAUACAAC----UAUUGAUAGACAUGGAUCGAUGUUUUUGGAUAGUGGGUGUGUUAUGCAAAUGCCACGCAUAAUAAACCCAGUAUAUCCGAAAUAAA-- ................----(((((((........))))))).(((((((((.(((((.((((((((.........)))))))).)))))...)))))))))....-- ( -28.90) >DroYak_CAF1 14438 106 - 1 CGAUACGGAAUACAUCUAUCUAUUGAUAGACAUGGAUCAAUGUUUUUGGAUAGUGGGUGUGUUAUGCAAAUGCCACGCAUAAAAGACCCAGUAUAUACGAAUUAAA-- .(((.((..((((..(((((((.....((((((......)))))).)))))))(((((.(.((((((.........)))))).).)))))))))...)).)))...-- ( -28.20) >consensus CGAUACAGAAUACAUC____UAUUGAUAGACAUGGAUCGAUGUUUUUGGAUAGUGGGUGUGUUAUGCAAAUGCCACGCAUAAUAAACCCAGUAUAUGCAAAAUAAA__ ....................(((((((........)))))))..((((((((.(((((.((((((((.........)))))))).)))))...))))))))....... (-21.06 = -22.50 + 1.44)


| Location | 3,661,620 – 3,661,731 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.97 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -26.90 |
| Energy contribution | -26.28 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3661620 111 + 22224390 CCAUGCCUAUCAAUA----GAUGUAUUCUGUAUCGCCGGAUGGAUCUUGUUAGGAGCACCUAGCUUGGAUUUAUGGCUAGUGCCAUAAUUAACUGGUCAAAGGUGUUGCUAAUGG ((((.((.....(((----((.....)))))......))))))...(..((((.(((((((.(((.(...(((((((....)))))))....).)))...))))))).))))..) ( -34.60) >DroSec_CAF1 13547 111 + 1 CCAUGCCUCUCAAUA----GAUGAAUUCUGUAUCGCCGGAUGGAUCUUGUUAGGAGCACCUAGCUUGGAUUUAUGGCCAGUGCCAUAAUUAACUGGUCGAAGGUGUUGCUAAUGA ((((.((.....(((----((.....)))))......))))))...(..((((.(((((((.(((.(...(((((((....)))))))....).)))...))))))).))))..) ( -33.40) >DroEre_CAF1 14427 111 + 1 CCAUGUCUAUCAAUA----GUUGUAUUCUGUAUCGCCGGAUAGAUCUUGUUAGGAGCACCCAGCUUCGAUUUAUGGCCGGUACCAUAAUUAACUGGUCGAAGGUGUUGCUAAUGG ((..(((((((....----((.((((...)))).))..)))))))...(((((.((((((....((((((((((((......)))))).......)))))))))))).))))))) ( -29.71) >DroYak_CAF1 14509 115 + 1 CCAUGUCUAUCAAUAGAUAGAUGUAUUCCGUAUCGCCGGAUAGAUCUCGUUAGGAGUAUCCAGCUUGGAUUUAUGGCUGGUGCCAUAAUUAACUAGUCGAAGGUGUUGCUAAUGG (((((((((((....))))))).......(((.(((((((((..(((.....))).))))).(((.(...(((((((....)))))))....).)))....)))).)))..)))) ( -35.10) >consensus CCAUGCCUAUCAAUA____GAUGUAUUCUGUAUCGCCGGAUAGAUCUUGUUAGGAGCACCCAGCUUGGAUUUAUGGCCAGUGCCAUAAUUAACUGGUCGAAGGUGUUGCUAAUGG .............................(((.((((..(((((((..((((((....))))))...)))))))(((((((..........)))))))...)))).)))...... (-26.90 = -26.28 + -0.62)


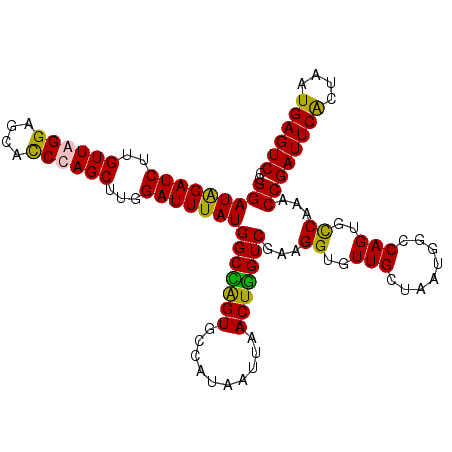
| Location | 3,661,651 – 3,661,760 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 92.81 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -30.01 |
| Energy contribution | -28.95 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3661651 109 + 22224390 CCGGAUGGAUCUUGUUAGGAGCACCUAGCUUGGAUUUAUGGCUAGUGCCAUAAUUAACUGGUCAAAGGUGUUGCUAAUGGCCAGUGCCAAACCGAUUCACUAAUGAGUC ..((.(((((((.((((((....))))))..))))(((((((....)))))))...((((((((..((.....))..)))))))).)))..))((((((....)))))) ( -37.40) >DroSec_CAF1 13578 109 + 1 CCGGAUGGAUCUUGUUAGGAGCACCUAGCUUGGAUUUAUGGCCAGUGCCAUAAUUAACUGGUCGAAGGUGUUGCUAAUGACCAGUGCCAAACCGAUUCACUAAUGAGUC ..((.(((((((.((((((....))))))..))))(((((((....)))))))...((((((((..((.....))..)))))))).)))..))((((((....)))))) ( -36.10) >DroEre_CAF1 14458 109 + 1 CCGGAUAGAUCUUGUUAGGAGCACCCAGCUUCGAUUUAUGGCCGGUACCAUAAUUAACUGGUCGAAGGUGUUGCUAAUGGCCAGUGCCAAACCGAUUCGCUAAUGAGUC ..((.........(((((.((((((....((((((((((((......)))))).......)))))))))))).)))))(((....)))...))((((((....)))))) ( -30.91) >DroYak_CAF1 14544 109 + 1 CCGGAUAGAUCUCGUUAGGAGUAUCCAGCUUGGAUUUAUGGCUGGUGCCAUAAUUAACUAGUCGAAGGUGUUGCUAAUGGCCAGUGUCAAACCGAUUCGCUAAUGAGUC ..((...(((((.((((..((((..((.(((.((((((((((....))))))).......))).))).)).))))..)))).)).)))...))((((((....)))))) ( -29.21) >consensus CCGGAUAGAUCUUGUUAGGAGCACCCAGCUUGGAUUUAUGGCCAGUGCCAUAAUUAACUGGUCGAAGGUGUUGCUAAUGGCCAGUGCCAAACCGAUUCACUAAUGAGUC ..(((((((((..((((((....))))))...)))))))(((((((..........)))))))...((..(((........)))..))...))((((((....)))))) (-30.01 = -28.95 + -1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:32 2006