
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,660,854 – 3,660,973 |
| Length | 119 |
| Max. P | 0.997904 |

| Location | 3,660,854 – 3,660,955 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.88 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -15.20 |
| Energy contribution | -18.36 |
| Covariance contribution | 3.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
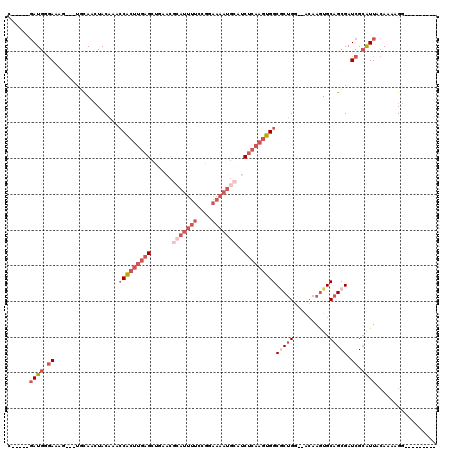
>X_DroMel_CAF1 3660854 101 + 22224390 ---------CCUUUUGUAAUGCGAUCGCUGCAUUUGU--CCAGCGCCACUUGAGAUGCAUUUUCCGGAAAAUGCGUUCAGCUCAAGUGGUUUGUAGUUGAA---CUUUCCCAUC-----G ---------..........(((((..((((.......--.))))(((((((((((((((((((....)))))))))....)))))))))))))))......---..........-----. ( -30.40) >DroSec_CAF1 12757 101 + 1 ---------CCUUUUGUAAUGCGAUCGCUGCACUUGU--CCAGUGCCACUUGAGAUGCAUUUUCCGGAAAAUGCGUUCAGCUCAAGUGGUUUGUAGUUGAA---CUUUCACAUC-----G ---------.........(((.((((((((((((...--..)))(((((((((((((((((((....)))))))))....))))))))))..)))).))).---...)).))).-----. ( -28.90) >DroSim_CAF1 17231 101 + 1 AAAUCGAACCCAAUUCAAACUCGAUCGCUGCACUUCC--GCUGGGCCAUU---GUUGCAUUUUCAA-----UUAGC-CCGCUCG-CCGCUUUGUAUUUACAAGGCAUUUACAU------- ..(((((.............))))).((.((......--)).((((....---((((......)))-----)..))-))))...-..(((((((....)))))))........------- ( -20.82) >DroEre_CAF1 13625 103 + 1 ---------CCUUUUGUAAUGCGAUCGCUGCACUUGUGCCCAGCGCCACUUGAGAUGCAUUUUCCGGAAAAUGCGUUAGGCUCAAGUGGCUUGUAGUUGCA---GUUUCCCAUC-----G ---------....(((((((((((..((((((....)))..)))(((((((((((((((((((....)))))))))....))))))))))))))).)))))---).........-----. ( -37.80) >DroYak_CAF1 13746 94 + 1 ---------CC---UGUAAUGCGAUCGCUGCACUUGU--UCAGCGCCACUUGAGAUGCAUUUUCCGGAAAA---------CUCAAGUGGCUUGUAGUUGCC---UUUUCCCAUCGUAUCG ---------..---....(((((((.((((.......--.))))((((((((((...(........)....---------))))))))))...........---.......))))))).. ( -26.50) >consensus _________CCUUUUGUAAUGCGAUCGCUGCACUUGU__CCAGCGCCACUUGAGAUGCAUUUUCCGGAAAAUGCGUUCAGCUCAAGUGGUUUGUAGUUGCA___CUUUCCCAUC_____G ..........................((((((...((.....))(((((((((((((((((((....)))))))))....)))))))))).))))))....................... (-15.20 = -18.36 + 3.16)



| Location | 3,660,854 – 3,660,955 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.88 |
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -13.22 |
| Energy contribution | -17.10 |
| Covariance contribution | 3.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
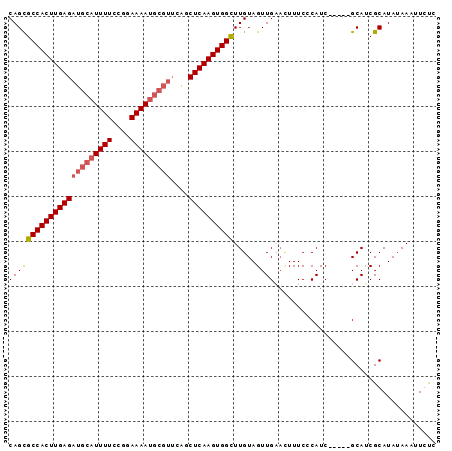
>X_DroMel_CAF1 3660854 101 - 22224390 C-----GAUGGGAAAG---UUCAACUACAAACCACUUGAGCUGAACGCAUUUUCCGGAAAAUGCAUCUCAAGUGGCGCUGG--ACAAAUGCAGCGAUCGCAUUACAAAAGG--------- .-----((((.((...---............(((((((((......(((((((....)))))))..)))))))))(((((.--.......))))).)).))))........--------- ( -30.90) >DroSec_CAF1 12757 101 - 1 C-----GAUGUGAAAG---UUCAACUACAAACCACUUGAGCUGAACGCAUUUUCCGGAAAAUGCAUCUCAAGUGGCACUGG--ACAAGUGCAGCGAUCGCAUUACAAAAGG--------- .-----(((((((..(---((...........((((((((......(((((((....)))))))..))))))))(((((..--...))))))))..)))))))........--------- ( -32.10) >DroSim_CAF1 17231 101 - 1 -------AUGUAAAUGCCUUGUAAAUACAAAGCGG-CGAGCGG-GCUAA-----UUGAAAAUGCAAC---AAUGGCCCAGC--GGAAGUGCAGCGAUCGAGUUUGAAUUGGGUUCGAUUU -------.......(((.((((....)))).)))(-(..((((-((((.-----(((.........)---)))))))).((--....)))).))(((((((..(.....)..))))))). ( -28.00) >DroEre_CAF1 13625 103 - 1 C-----GAUGGGAAAC---UGCAACUACAAGCCACUUGAGCCUAACGCAUUUUCCGGAAAAUGCAUCUCAAGUGGCGCUGGGCACAAGUGCAGCGAUCGCAUUACAAAAGG--------- .-----((((.((...---...........((((((((((......(((((((....)))))))..))))))))))((((.((....)).))))..)).))))........--------- ( -37.20) >DroYak_CAF1 13746 94 - 1 CGAUACGAUGGGAAAA---GGCAACUACAAGCCACUUGAG---------UUUUCCGGAAAAUGCAUCUCAAGUGGCGCUGA--ACAAGUGCAGCGAUCGCAUUACA---GG--------- ......((((.((..(---(....))....((((((((((---------.................))))))))))((((.--.......))))..)).))))...---..--------- ( -28.83) >consensus C_____GAUGGGAAAG___UGCAACUACAAACCACUUGAGCUGAACGCAUUUUCCGGAAAAUGCAUCUCAAGUGGCGCUGG__ACAAGUGCAGCGAUCGCAUUACAAAAGG_________ ......((((.((..................(((((((((......(((((((....)))))))..)))))))))(((((..........))))).)).))))................. (-13.22 = -17.10 + 3.88)

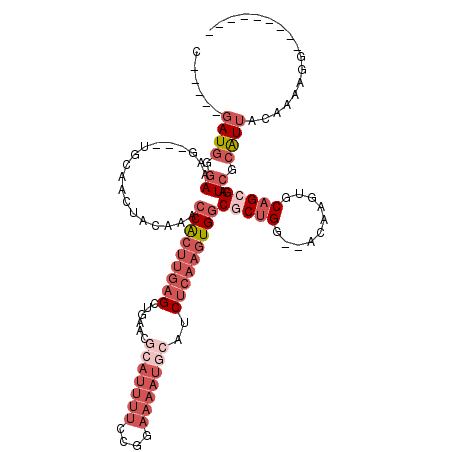
| Location | 3,660,883 – 3,660,973 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 85.41 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -21.21 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3660883 90 + 22224390 CAGCGCCACUUGAGAUGCAUUUUCCGGAAAAUGCGUUCAGCUCAAGUGGUUUGUAGUUGAACUUUCCCAUC-----GCAUCGCAUAGAAAUUCUC ..(((((((((((((((((((((....)))))))))....)))))))))).(((...((........))..-----)))..))............ ( -25.80) >DroSec_CAF1 12786 90 + 1 CAGUGCCACUUGAGAUGCAUUUUCCGGAAAAUGCGUUCAGCUCAAGUGGUUUGUAGUUGAACUUUCACAUC-----GCAUCGCAUAUAAAUUCUC ..(((((((((((((((((((((....)))))))))....)))))))))).((..(((((....))).)).-----.))..))............ ( -25.00) >DroEre_CAF1 13656 90 + 1 CAGCGCCACUUGAGAUGCAUUUUCCGGAAAAUGCGUUAGGCUCAAGUGGCUUGUAGUUGCAGUUUCCCAUC-----GCAGCGCAUAUAAAUUGCC ..(((((((((((((((((((((....)))))))))....)))))))))).....(((((...........-----)))))))............ ( -33.80) >DroYak_CAF1 13772 86 + 1 CAGCGCCACUUGAGAUGCAUUUUCCGGAAAA---------CUCAAGUGGCUUGUAGUUGCCUUUUCCCAUCGUAUCGCAUCGCAUAUAAAUUGUC ..((((((((((((...(........)....---------)))))))))).....(.(((................))).)))............ ( -19.29) >consensus CAGCGCCACUUGAGAUGCAUUUUCCGGAAAAUGCGUUCAGCUCAAGUGGCUUGUAGUUGAACUUUCCCAUC_____GCAUCGCAUAUAAAUUCUC (((((((((((((((((((((((....)))))))))....)))))))))).....)))).................................... (-21.21 = -22.03 + 0.81)


| Location | 3,660,883 – 3,660,973 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 85.41 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -17.08 |
| Energy contribution | -18.32 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
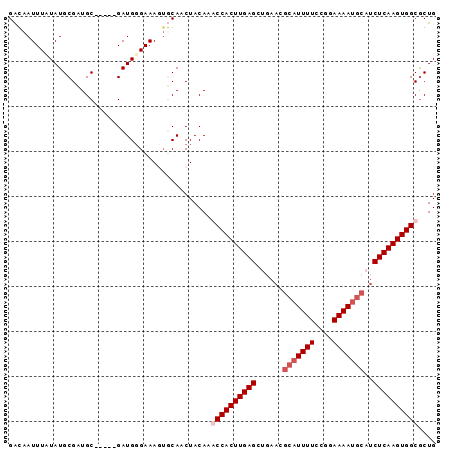
>X_DroMel_CAF1 3660883 90 - 22224390 GAGAAUUUCUAUGCGAUGC-----GAUGGGAAAGUUCAACUACAAACCACUUGAGCUGAACGCAUUUUCCGGAAAAUGCAUCUCAAGUGGCGCUG (((..(((((((((...))-----.)))))))..))).........(((((((((......(((((((....)))))))..)))))))))..... ( -25.80) >DroSec_CAF1 12786 90 - 1 GAGAAUUUAUAUGCGAUGC-----GAUGUGAAAGUUCAACUACAAACCACUUGAGCUGAACGCAUUUUCCGGAAAAUGCAUCUCAAGUGGCACUG (((..(((((((((...))-----.)))))))..))).........(((((((((......(((((((....)))))))..)))))))))..... ( -24.60) >DroEre_CAF1 13656 90 - 1 GGCAAUUUAUAUGCGCUGC-----GAUGGGAAACUGCAACUACAAGCCACUUGAGCCUAACGCAUUUUCCGGAAAAUGCAUCUCAAGUGGCGCUG .(((.......)))(((((-----...((....))))).......((((((((((......(((((((....)))))))..)))))))))))).. ( -30.60) >DroYak_CAF1 13772 86 - 1 GACAAUUUAUAUGCGAUGCGAUACGAUGGGAAAAGGCAACUACAAGCCACUUGAG---------UUUUCCGGAAAAUGCAUCUCAAGUGGCGCUG .................((..............((....))....((((((((((---------.................)))))))))))).. ( -20.83) >consensus GACAAUUUAUAUGCGAUGC_____GAUGGGAAAGUGCAACUACAAACCACUUGAGCUGAACGCAUUUUCCGGAAAAUGCAUCUCAAGUGGCGCUG .............................................((((((((((......(((((((....)))))))..)))))))))).... (-17.08 = -18.32 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:29 2006