| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,657,664 – 3,657,794 |
| Length | 130 |
| Max. P | 0.883189 |

| Location | 3,657,664 – 3,657,764 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.48 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
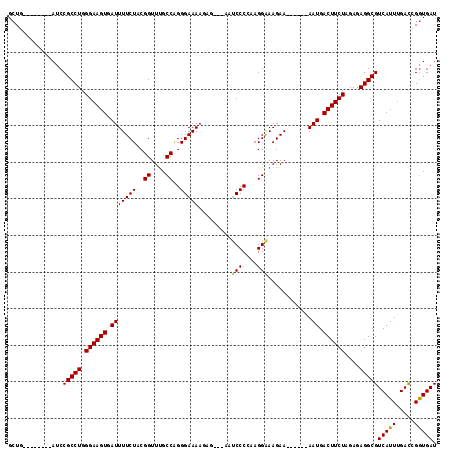
>X_DroMel_CAF1 3657664 100 + 22224390 GCUG--------AUCCGCCUGGGAAGUGAUUUUCUACGGUUUGCCAGGGAAAAGAG---AAUCCCCAAGGAAAGAA------AAUGACUUCUAGACAGGCGUCAUUUGACCGGUGAU ((((--------....(((((((((((.(((((((..((....)).((((......---..)))).......))))------))).))))))...)))))(((....)))))))... ( -35.00) >DroSec_CAF1 9581 100 + 1 GCUG--------AUCCGCCUGGGAAGUGAUUUUCUACGGUUCGCCAGGGAAAAGAG---AAUCCCCAAGGAAAGAA------AAUGACUUCUAGAGAGGCGUCAUUUGACCGGUGAU ((((--------....((((.((((((.(((((((..((....)).((((......---..)))).......))))------))).))))))....))))(((....)))))))... ( -33.00) >DroSim_CAF1 5413 100 + 1 GCUG--------AUCCGCCUGGGAAGUGAUUUUCUACGGUUUGCCGGGGAAAAGAG---AAUCCCCAAGGAAAGAA------AAUGACUUCUAGAGAGGCGUCAUUUGACCGGUGAU ((((--------....((((.((((((.(((((((..((....))(((((......---..)))))......))))------))).))))))....))))(((....)))))))... ( -37.30) >DroEre_CAF1 10546 114 + 1 GCUGAUCCGCUGAUCCGCCUGGGAAGUGAUUUUCUACGGUUUGCCAGGGAAAACAG---AACCCCGAAGGGAAGAAAAUGAAAAUGACUUCUGGAGAGGCGUCAUUUGAUAGGUGAU (((..(((((......))...((((((.((((((.....(((.((.(((.......---...)))...)).))).....)))))).)))))))))..)))((((((.....)))))) ( -33.90) >DroYak_CAF1 10599 102 + 1 GCUG--------AUCCGCCUGGGAAGUGAUUUUCUACGGUUUGCCAGGGAAA-CAGGGAAACCCCAAAGGAAAGAA------AAUGACUUCUGGAGAGGCGUCAUUGGACGGAUGAU ....--------((((((((.((((((.(((((((..(((((.((..(....-).)).)))))((...))..))))------))).))))))....))))(((....)))))))... ( -37.50) >consensus GCUG________AUCCGCCUGGGAAGUGAUUUUCUACGGUUUGCCAGGGAAAAGAG___AAUCCCCAAGGAAAGAA______AAUGACUUCUAGAGAGGCGUCAUUUGACCGGUGAU ...............(((((.((((((.(((((((..((....))..))))).........(((....)))............)).))))))....)))))(((((.....))))). (-22.68 = -22.48 + -0.20)


| Location | 3,657,664 – 3,657,764 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.24 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3657664 100 - 22224390 AUCACCGGUCAAAUGACGCCUGUCUAGAAGUCAUU------UUCUUUCCUUGGGGAUU---CUCUUUUCCCUGGCAAACCGUAGAAAAUCACUUCCCAGGCGGAU--------CAGC ....((.(((....)))(((((....(((((.(((------((((......(((((..---......)))))((....))..))))))).))))).)))))))..--------.... ( -31.30) >DroSec_CAF1 9581 100 - 1 AUCACCGGUCAAAUGACGCCUCUCUAGAAGUCAUU------UUCUUUCCUUGGGGAUU---CUCUUUUCCCUGGCGAACCGUAGAAAAUCACUUCCCAGGCGGAU--------CAGC ....((.(((....)))((((.....(((((.(((------(((((((.(..((((..---......))))..).)))....))))))).)))))..))))))..--------.... ( -31.00) >DroSim_CAF1 5413 100 - 1 AUCACCGGUCAAAUGACGCCUCUCUAGAAGUCAUU------UUCUUUCCUUGGGGAUU---CUCUUUUCCCCGGCAAACCGUAGAAAAUCACUUCCCAGGCGGAU--------CAGC ....((.(((....)))((((.....(((((.(((------((((......(((((..---......)))))((....))..))))))).)))))..))))))..--------.... ( -30.90) >DroEre_CAF1 10546 114 - 1 AUCACCUAUCAAAUGACGCCUCUCCAGAAGUCAUUUUCAUUUUCUUCCCUUCGGGGUU---CUGUUUUCCCUGGCAAACCGUAGAAAAUCACUUCCCAGGCGGAUCAGCGGAUCAGC ................(((((.....(((((.((((((........(((....)))..---..((((.(....).))))....)))))).)))))..)))))((((....))))... ( -28.90) >DroYak_CAF1 10599 102 - 1 AUCAUCCGUCCAAUGACGCCUCUCCAGAAGUCAUU------UUCUUUCCUUUGGGGUUUCCCUG-UUUCCCUGGCAAACCGUAGAAAAUCACUUCCCAGGCGGAU--------CAGC ...(((((((....)))((((.....(((((.(((------((((..((...))(((((.((..-.......)).)))))..))))))).)))))..))))))))--------.... ( -27.50) >consensus AUCACCGGUCAAAUGACGCCUCUCUAGAAGUCAUU______UUCUUUCCUUGGGGAUU___CUCUUUUCCCUGGCAAACCGUAGAAAAUCACUUCCCAGGCGGAU________CAGC ....((.(((....)))((((.....(((((...............(((....)))........(((((..(((....)))..)))))..)))))..)))))).............. (-19.56 = -19.24 + -0.32)


| Location | 3,657,693 – 3,657,794 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.94 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3657693 101 + 22224390 GGUUUGCCAGGGAAAAGAG---AAUCCCCAAGGAAAGAA------AAUGACUUCUAGACAGGCGUCAUUUGACCGGUGAUCAGGGUUU--------CCCGUCAAAGGUGAAACAAAUU-- ((.(..((.((((......---..))))...((.....(------((((((.(((....))).)))))))..))))..)))...((((--------(((......)).))))).....-- ( -26.50) >DroSec_CAF1 9610 103 + 1 GGUUCGCCAGGGAAAAGAG---AAUCCCCAAGGAAAGAA------AAUGACUUCUAGAGAGGCGUCAUUUGACCGGUGAUCAGGGUUU--------CCCGUCAAAGGUGAAACAAAUUUU (((.((((.((((......---..))))...((.....(------((((((.(((....))).)))))))..)))))))))...((((--------(((......)).)))))....... ( -29.20) >DroSim_CAF1 5442 103 + 1 GGUUUGCCGGGGAAAAGAG---AAUCCCCAAGGAAAGAA------AAUGACUUCUAGAGAGGCGUCAUUUGACCGGUGAUCAGGGUUU--------CCCGUCAAAGGUGAAACAAAUUUU ((.(..(((((((......---..)))))..((.....(------((((((.(((....))).)))))))..))))..)))...((((--------(((......)).)))))....... ( -31.10) >DroEre_CAF1 10583 109 + 1 GGUUUGCCAGGGAAAACAG---AACCCCGAAGGGAAGAAAAUGAAAAUGACUUCUGGAGAGGCGUCAUUUGAUAGGUGAUCAGGGUUU--------CCCGUCAAAGGUGAAACAAAUUUU .(((((((.(((.......---...)))((.((((((....((((((((((.(((....))).))))))).........)))...)))--------))).))...))).))))....... ( -27.30) >DroYak_CAF1 10628 103 + 1 GGUUUGCCAGGGAAA-CAGGGAAACCCCAAAGGAAAGAA------AAUGACUUCUGGAGAGGCGUCAUUGGACGGAUGAUCAGGGUUUCCCGGUUUCCCGUCAAAGGUGA---------- ((....)).((((((-(.(((((((((............------.....((((....))))((((....))))........))))))))).)))))))...........---------- ( -38.50) >consensus GGUUUGCCAGGGAAAAGAG___AAUCCCCAAGGAAAGAA______AAUGACUUCUAGAGAGGCGUCAUUUGACCGGUGAUCAGGGUUU________CCCGUCAAAGGUGAAACAAAUUUU ..((((((.(((.............)))...................((((.....((..(.((((....))).).)..)).(((...........)))))))..))))))......... (-18.82 = -18.70 + -0.12)



| Location | 3,657,693 – 3,657,794 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.94 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -15.34 |
| Energy contribution | -14.66 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3657693 101 - 22224390 --AAUUUGUUUCACCUUUGACGGG--------AAACCCUGAUCACCGGUCAAAUGACGCCUGUCUAGAAGUCAUU------UUCUUUCCUUGGGGAUU---CUCUUUUCCCUGGCAAACC --...(..(((.(((..(((((((--------....)))).)))..))).)))..).(((.....(((((....)------))))......(((((..---......))))))))..... ( -25.70) >DroSec_CAF1 9610 103 - 1 AAAAUUUGUUUCACCUUUGACGGG--------AAACCCUGAUCACCGGUCAAAUGACGCCUCUCUAGAAGUCAUU------UUCUUUCCUUGGGGAUU---CUCUUUUCCCUGGCGAACC .....(..(((.(((..(((((((--------....)))).)))..))).)))..)((((.....(((((....)------))))......(((((..---......))))))))).... ( -27.70) >DroSim_CAF1 5442 103 - 1 AAAAUUUGUUUCACCUUUGACGGG--------AAACCCUGAUCACCGGUCAAAUGACGCCUCUCUAGAAGUCAUU------UUCUUUCCUUGGGGAUU---CUCUUUUCCCCGGCAAACC .....(..(((.(((..(((((((--------....)))).)))..))).)))..).(((.....(((((....)------))))......(((((..---......))))))))..... ( -28.50) >DroEre_CAF1 10583 109 - 1 AAAAUUUGUUUCACCUUUGACGGG--------AAACCCUGAUCACCUAUCAAAUGACGCCUCUCCAGAAGUCAUUUUCAUUUUCUUCCCUUCGGGGUU---CUGUUUUCCCUGGCAAACC ....((((((........((((((--------..(((((((.........(((((((...((....)).)))))))..............))))))))---)))))......)))))).. ( -21.94) >DroYak_CAF1 10628 103 - 1 ----------UCACCUUUGACGGGAAACCGGGAAACCCUGAUCAUCCGUCCAAUGACGCCUCUCCAGAAGUCAUU------UUCUUUCCUUUGGGGUUUCCCUG-UUUCCCUGGCAAACC ----------.....((((..(((((((.(((((((((..(..........((((((...((....)).))))))------.........)..))))))))).)-))))))...)))).. ( -39.01) >consensus AAAAUUUGUUUCACCUUUGACGGG________AAACCCUGAUCACCGGUCAAAUGACGCCUCUCUAGAAGUCAUU______UUCUUUCCUUGGGGAUU___CUCUUUUCCCUGGCAAACC ...............((((.((((..............((((.....))))((((((..((....))..))))))...........(((....))).............)))).)))).. (-15.34 = -14.66 + -0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:24 2006