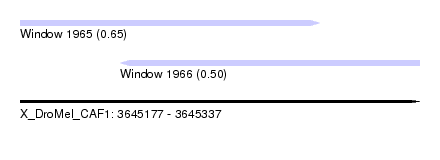
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,645,177 – 3,645,337 |
| Length | 160 |
| Max. P | 0.654470 |

| Location | 3,645,177 – 3,645,297 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -42.76 |
| Consensus MFE | -32.24 |
| Energy contribution | -32.48 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3645177 120 + 22224390 CGGCCUCCGCUUACGGCCAGCAGAAGUUCGACGAGGCCAUCGCCUUGUACACCAAGGCCAUUGAACUGAGCCCGGGUAAUGCCCUUUUCCACGCUAAGCGUGGUCAGGCCUUCCUCAAGC .(((((..(((((.((((.(..(.....)..)..))))...((((((.....))))))........)))))..(((.....)))....((((((...))))))..))))).......... ( -42.20) >DroSec_CAF1 15150 120 + 1 CGGCCUCCGCCUACGGCGAGCAGAAGUUCGACGAGGCCAUCGCCUUGUACACCAAGGCCAUUGAACUGAGCCCGGGUAAUGCCCUUUUCCACGCUAAGCGUGGUCAGGCAUUCCUCAAGC (((.(((.((((.((.(((((....))))).))))))((..((((((.....))))))...))....))).)))((.((((((.....((((((...))))))...))))))))...... ( -49.00) >DroSim_CAF1 7142 120 + 1 CGGCCUCCGCCUACGGCCAGCAGAAGUUCGACGAGGCCAUCGCCUUGUACACCAAGGCCAUUGAACUGAGCCCGGGUAAUGCCCUUUUCCACGCUAAGCGUGGUCAGGCCUUCCUCAAGC .(((....)))...((((.(((........(((((((....)))))))..(((..(((((......)).)))..)))..)))......((((((...))))))...)))).......... ( -41.80) >DroEre_CAF1 16076 120 + 1 CGGCCUCUGCCUACGGCCAGCAGAAGUUCGACGAGGCAAUCGCCUUCUACACCAAGGCCAUCGAACUAAACCCGGGCAAUGCCCUGUUCCACGCCAAGCGUGGCCAGGCCUUUCUCAAGC .((((((((((....)...)))))(((((((.((.....))(((((.......)))))..)))))))......((((...))))....((((((...))))))...)))).......... ( -40.90) >DroWil_CAF1 8875 120 + 1 CAGCUGCCGCCUACAGUGAGCAGAAAUUCGAUGAGGCCAUCGAAUUCUAUACCAAGGCCAUUGAGCUCAAUCCGGGCAAUGCUCUGUUCUACGCCAAGCGUGGUCAGGCUUUCCUUAAGC ..(((...((((....(((((((((.(((((((....)))))))))))....(((.....))).)))))....))))...((.(((..((((((...)))))).)))))........))) ( -39.90) >consensus CGGCCUCCGCCUACGGCCAGCAGAAGUUCGACGAGGCCAUCGCCUUGUACACCAAGGCCAUUGAACUGAGCCCGGGUAAUGCCCUUUUCCACGCUAAGCGUGGUCAGGCCUUCCUCAAGC .((((.........)))).((.........(((((((....))))))).....((((((..............((((...))))....((((((...))))))...))))))......)) (-32.24 = -32.48 + 0.24)


| Location | 3,645,217 – 3,645,337 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.33 |
| Mean single sequence MFE | -45.54 |
| Consensus MFE | -36.64 |
| Energy contribution | -37.56 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3645217 120 - 22224390 GCCACGUCGCAGUCCCGAAUGCAGGCAUUCGGCUUCUUCAGCUUGAGGAAGGCCUGACCACGCUUAGCGUGGAAAAGGGCAUUACCCGGGCUCAGUUCAAUGGCCUUGGUGUACAAGGCG (((...........(((((((....)))))))(((((((.....)))))))((((..((((((...))))))....))))..((((.((((.((......)))))).)))).....))). ( -50.20) >DroSec_CAF1 15190 120 - 1 GCCAUGUCGCAGUCCCGAAUGCAGGCAUUCGGCUUCUUCAGCUUGAGGAAUGCCUGACCACGCUUAGCGUGGAAAAGGGCAUUACCCGGGCUCAGUUCAAUGGCCUUGGUGUACAAGGCG ..(((...((....(((((((....))))))).......((((((.(((((((((..((((((...))))))....))))))).))))))))..))...)))((((((.....)))))). ( -51.10) >DroSim_CAF1 7182 120 - 1 GCCAUGUCGCAGUCCCGAAUGCAGGCAUUCGGCUUCUUCAGCUUGAGGAAGGCCUGACCACGCUUAGCGUGGAAAAGGGCAUUACCCGGGCUCAGUUCAAUGGCCUUGGUGUACAAGGCG (((.(((.((....(((((((....)))))))(((((((.....)))))))((((..((((((...))))))....))))...(((.((((.((......)))))).)))))))).))). ( -52.00) >DroEre_CAF1 16116 120 - 1 GCCUUGUCGCAGUCCCGAAUGCAGGCAUUCGGUUUCUUCAGCUUGAGAAAGGCCUGGCCACGCUUGGCGUGGAACAGGGCAUUGCCCGGGUUUAGUUCGAUGGCCUUGGUGUAGAAGGCG (((((.((((....(((((((....)))))))((((........))))((((((...((((((...))))))(((.((((...))))..))).........)))))).))).).))))). ( -48.20) >DroWil_CAF1 8915 120 - 1 GCCUUAUCGCAGUCGCGUAUGCAUGCAUUUGGUUUCUUAAGCUUAAGGAAAGCCUGACCACGCUUGGCGUAGAACAGAGCAUUGCCCGGAUUGAGCUCAAUGGCCUUGGUAUAGAAUUCG ((..((.(((....))))).))((((....((((.....(((((((((((.(((((..(((((...)))).)..))).)).)).))....)))))))....))))...))))........ ( -26.20) >consensus GCCAUGUCGCAGUCCCGAAUGCAGGCAUUCGGCUUCUUCAGCUUGAGGAAGGCCUGACCACGCUUAGCGUGGAAAAGGGCAUUACCCGGGCUCAGUUCAAUGGCCUUGGUGUACAAGGCG (((...........(((((((....)))))))(((((((.....)))))))((((..((((((...))))))....))))..((((.(((((.........))))).)))).....))). (-36.64 = -37.56 + 0.92)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:18 2006