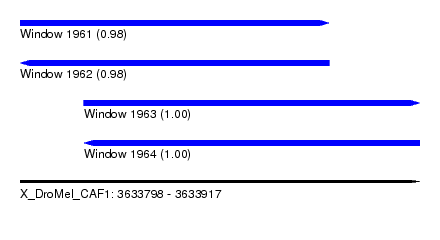
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,633,798 – 3,633,917 |
| Length | 119 |
| Max. P | 0.998508 |

| Location | 3,633,798 – 3,633,890 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 98.70 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -24.10 |
| Energy contribution | -23.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3633798 92 + 22224390 GUAACCGUUAGAAUUAGCCCAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGA ...(((((((...........)))))))((.(((((((.(((...((...(((((((......)))))))...))...))).))))))))). ( -24.10) >DroSec_CAF1 12855 92 + 1 GUAACCGUUAGAAAUAGCCCAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGA ...(((((((...........)))))))((.(((((((.(((...((...(((((((......)))))))...))...))).))))))))). ( -24.10) >DroSim_CAF1 4625 92 + 1 GUAACCGUUAGAAUUAGCCCAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGA ...(((((((...........)))))))((.(((((((.(((...((...(((((((......)))))))...))...))).))))))))). ( -24.10) >DroEre_CAF1 13536 92 + 1 GUAACCGUUAGAAUUAGCCCAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAGCGAUUGGAUGCGA ...(((((((...........)))))))((.(((((((.(((...((...(((((((......)))))))...))...))).))))))))). ( -24.40) >DroYak_CAF1 5180 92 + 1 GUAACCGUUAGAAUUAGCCCAUAAUGGUCGAUAUCUAAAUGUGUAUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGA ...(((((((...........)))))))((.(((((((.(((...((...(((((((......)))))))...))...))).))))))))). ( -23.00) >consensus GUAACCGUUAGAAUUAGCCCAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGA ...(((((((...........)))))))((.(((((((.(((...((...(((((((......)))))))...))...))).))))))))). (-24.10 = -23.94 + -0.16)


| Location | 3,633,798 – 3,633,890 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 98.70 |
| Mean single sequence MFE | -16.90 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.44 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3633798 92 - 22224390 UCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUGGGCUAAUUCUAACGGUUAC ..((..(((...(((((((.....)))))))......(((((((..............)))))))......)))))................ ( -17.14) >DroSec_CAF1 12855 92 - 1 UCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUGGGCUAUUUCUAACGGUUAC ..((..(((...(((((((.....)))))))......(((((((..............)))))))......)))))................ ( -17.14) >DroSim_CAF1 4625 92 - 1 UCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUGGGCUAAUUCUAACGGUUAC ..((..(((...(((((((.....)))))))......(((((((..............)))))))......)))))................ ( -17.14) >DroEre_CAF1 13536 92 - 1 UCGCAUCCAAUCGCUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUGGGCUAAUUCUAACGGUUAC ........(((((.........(((((((......)))))))............(((((......((.....))......)))))))))).. ( -15.50) >DroYak_CAF1 5180 92 - 1 UCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACAUACACAUUUAGAUAUCGACCAUUAUGGGCUAAUUCUAACGGUUAC ..((..(((...(((((((.....)))))))......(((((((...((......)).)))))))......)))))................ ( -17.60) >consensus UCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUGGGCUAAUUCUAACGGUUAC ..((..(((...(((((((.....)))))))......(((((((..............)))))))......)))))................ (-16.24 = -16.44 + 0.20)


| Location | 3,633,817 – 3,633,917 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -23.42 |
| Energy contribution | -23.78 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3633817 100 + 22224390 CAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGAGUGUGUGUUAAUUUAAUUUGAUAAAAG ....(((.(((.(((((((.(((...((...(((((((......)))))))...))...))).)))))))))).))).((((((......)))))).... ( -24.20) >DroSec_CAF1 12874 100 + 1 CAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGAGAGUGUGUUAAUUUAAUUUGAUAAAAG ....((..(((.(((((((.(((...((...(((((((......)))))))...))...))).))))))))))..)).((((((......)))))).... ( -23.10) >DroSim_CAF1 4644 100 + 1 CAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGAGAGUGUGUUAAUUUAAUUUGAUAAAAG ....((..(((.(((((((.(((...((...(((((((......)))))))...))...))).))))))))))..)).((((((......)))))).... ( -23.10) >DroEre_CAF1 13555 100 + 1 CAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAGCGAUUGGAUGCGAGCGUGUGUUAAUUUAAUUUGAUAAAAG ....(((.(((.(((((((.(((...((...(((((((......)))))))...))...))).)))))))))).))).((((((......)))))).... ( -26.80) >DroYak_CAF1 5199 100 + 1 CAUAAUGGUCGAUAUCUAAAUGUGUAUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGAGCGUGUGUUAAUUUAAUUUCAUAAAAG ((((.((.(((.(((((((.(((...((...(((((((......)))))))...))...))).)))))))))).)))))).................... ( -24.40) >consensus CAUAAUGGUCGAUAUCUAAAUGUGUGUGUAUAUCGAUGUGAUGUCAUCGAUUGUCGAUAACGAUUGGAUGCGAGAGUGUGUUAAUUUAAUUUGAUAAAAG ....(((.(((.(((((((.(((...((...(((((((......)))))))...))...))).)))))))))).))).((((((......)))))).... (-23.42 = -23.78 + 0.36)



| Location | 3,633,817 – 3,633,917 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -13.30 |
| Consensus MFE | -12.74 |
| Energy contribution | -12.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3633817 100 - 22224390 CUUUUAUCAAAUUAAAUUAACACACACUCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUG .......................................(((((((.....)))))))......(((((((..............)))))))........ ( -13.64) >DroSec_CAF1 12874 100 - 1 CUUUUAUCAAAUUAAAUUAACACACUCUCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUG .......................................(((((((.....)))))))......(((((((..............)))))))........ ( -13.64) >DroSim_CAF1 4644 100 - 1 CUUUUAUCAAAUUAAAUUAACACACUCUCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUG .......................................(((((((.....)))))))......(((((((..............)))))))........ ( -13.64) >DroEre_CAF1 13555 100 - 1 CUUUUAUCAAAUUAAAUUAACACACGCUCGCAUCCAAUCGCUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUG ....((((................((...((........))...))...(((((((......)))))))...............))))............ ( -11.40) >DroYak_CAF1 5199 100 - 1 CUUUUAUGAAAUUAAAUUAACACACGCUCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACAUACACAUUUAGAUAUCGACCAUUAUG ....(((((..............................(((((((.....)))))))......(((((((...((......)).)))))))...))))) ( -14.20) >consensus CUUUUAUCAAAUUAAAUUAACACACGCUCGCAUCCAAUCGUUAUCGACAAUCGAUGACAUCACAUCGAUAUACACACACAUUUAGAUAUCGACCAUUAUG .......................................(((((((.....)))))))......(((((((..............)))))))........ (-12.74 = -12.94 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:16 2006