
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,622,714 – 3,622,809 |
| Length | 95 |
| Max. P | 0.559518 |

| Location | 3,622,714 – 3,622,809 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -19.71 |
| Energy contribution | -21.52 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
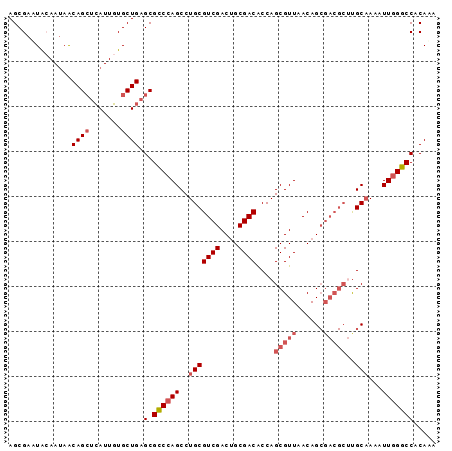
>X_DroMel_CAF1 3622714 95 + 22224390 AGCGAAUAGAAUAACAGCUCAUUGUGCUGAGCGCCCAGCCUGCGUCGACUGCGACACCAGCGUUAACAGCGACGCCUGCAAAAUUGGGCCACAAA ................(((((......)))))(((((((..((((((.(((.(((......)))..)))))))))..)).....)))))...... ( -30.60) >DroSec_CAF1 1605 95 + 1 AGCGAAUACAAUAACAGCUCAUUGUGCUGAGCGUCCAGCCUGCGUCGACUGCGACACCAGCGUUAACAGCGACGCUUGCAAAAUUGGGCCACAAA ................(((((......)))))(((((((..((((((.(((.(((......)))..)))))))))..)).....)))))...... ( -28.80) >DroEre_CAF1 1598 95 + 1 AGCGAUUACAAUAACAGCUCAUUGUGCUGAGCGCCCAGCCUGCGUCGACUGCGACACCAGCGUUAACAGCGACGCUCGCAAAAUUGGGCCACAAA ................(((((......)))))(((((((..((((((.(((.(((......)))..)))))))))..)).....)))))...... ( -31.40) >DroAna_CAF1 6771 80 + 1 AGCGAGUG-----GCAGG-GA--UUGCUGCGCGCCGAGCCUGCGUCGACAGCGACGCCAGCGUUAACAG-------UGCCAAAUUGGGCCACACA .....(((-----((...-((--((...((((....(((((((((((....))))).))).)))....)-------)))..))))..)))))... ( -30.40) >consensus AGCGAAUACAAUAACAGCUCAUUGUGCUGAGCGCCCAGCCUGCGUCGACUGCGACACCAGCGUUAACAGCGACGCUUGCAAAAUUGGGCCACAAA ..............((((.......)))).(.((((((..(((((((....))))....(((((......)))))..)))...)))))))..... (-19.71 = -21.52 + 1.81)


| Location | 3,622,714 – 3,622,809 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 83.68 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -23.42 |
| Energy contribution | -24.30 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
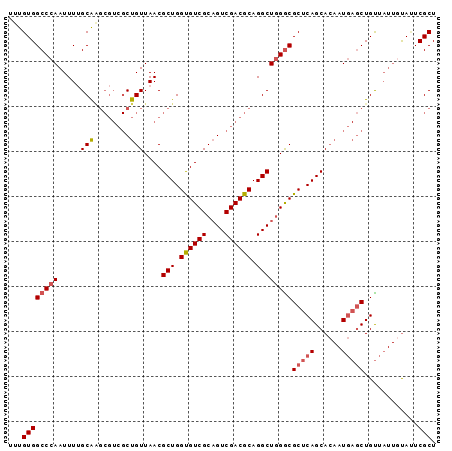
>X_DroMel_CAF1 3622714 95 - 22224390 UUUGUGGCCCAAUUUUGCAGGCGUCGCUGUUAACGCUGGUGUCGCAGUCGACGCAGGCUGGGCGCUCAGCACAAUGAGCUGUUAUUCUAUUCGCU ...((((((((.....((..((((((((((..((....))...)))).))))))..)))))))(((((......)))))............))). ( -37.20) >DroSec_CAF1 1605 95 - 1 UUUGUGGCCCAAUUUUGCAAGCGUCGCUGUUAACGCUGGUGUCGCAGUCGACGCAGGCUGGACGCUCAGCACAAUGAGCUGUUAUUGUAUUCGCU ...((((..((((...(((.((.(((.(((....(((((((((.(((((......)))))))))).))))))).)))))))).))))...)))). ( -32.00) >DroEre_CAF1 1598 95 - 1 UUUGUGGCCCAAUUUUGCGAGCGUCGCUGUUAACGCUGGUGUCGCAGUCGACGCAGGCUGGGCGCUCAGCACAAUGAGCUGUUAUUGUAAUCGCU ...((((((((.....((..((((((((((..((....))...)))).))))))..)))))))(((((......)))))............))). ( -35.50) >DroAna_CAF1 6771 80 - 1 UGUGUGGCCCAAUUUGGCA-------CUGUUAACGCUGGCGUCGCUGUCGACGCAGGCUCGGCGCGCAGCAA--UC-CCUGC-----CACUCGCU .(.(((((((.....))..-------........((((((((((..(((......))).)))))).))))..--..-...))-----))).)... ( -30.00) >consensus UUUGUGGCCCAAUUUUGCAAGCGUCGCUGUUAACGCUGGUGUCGCAGUCGACGCAGGCUGGGCGCUCAGCACAAUGAGCUGUUAUUGUAUUCGCU ...((((((((.....(((........)))....(((.((((((....)))))).))))))))(((((......)))))............))). (-23.42 = -24.30 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:04:07 2006