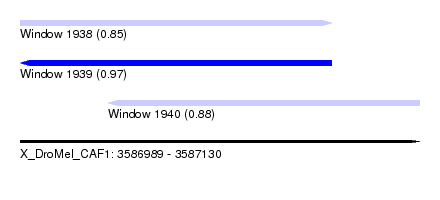
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,586,989 – 3,587,130 |
| Length | 141 |
| Max. P | 0.968414 |

| Location | 3,586,989 – 3,587,099 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.88 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -13.02 |
| Energy contribution | -16.86 |
| Covariance contribution | 3.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
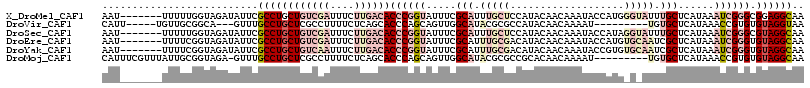
>X_DroMel_CAF1 3586989 110 + 22224390 AAU-------UUUUUGGUAGAUAUUCGCCUGCUGUCGAUUUCUUGACACCCGGUAUUUCGCAUUUGCUCCAUACAACAAAUACCAUGGGUAUUUGCUCAUAAAUCGGGCGGAGGCAA ...-------................((((.(((((((....))))))((((((.....(((..(((.((((............)))))))..)))......)))))).).)))).. ( -29.90) >DroVir_CAF1 76802 100 + 1 CAUU-----UGUUGCGGCA---GUUUGCCUGCUCGCCUUUUCUCAGCACCCAGCAGUUGGCAUACGCGCCAUACAACAAAAU---------UGUGCUCAUAAACCGUGUGUAGGUAA ..((-----(((((.(((.---((.((((.(((.((................))))).)))).))..)))...)))))))..---------(.(((.(((.....))).))).)... ( -24.49) >DroSec_CAF1 45965 110 + 1 AAU-------UUUUUGGUAGAUAUUCGCCUGCUGUCGAUUUCUUGACACCCGGUAUUUCGCAUUUGCUCCAUACAACAAAUACCAUAGGUAUUUGCUCAUAAAUCGGGCGUAGGCAA ...-------................((((((((((((....))))))((((((((...((....))...))))..((((((((...)))))))).........)))).)))))).. ( -32.30) >DroEre_CAF1 40776 110 + 1 AAU-------UUUUCGGUAGAUAUUCGCCUGCUGUCGAUUUCUUGACACCCGGUAUUUCGCAUUUGCGACAUACAACAAAUACCAUGUGCAAUCGCUCAUAAAUCGGGUGUAGGCAA ...-------................((((((.(((((....)))))(((((((((.((((....)))).))))..........(((.((....)).)))....))))))))))).. ( -35.40) >DroYak_CAF1 41684 110 + 1 AAU-------UUUUCGGUAGAUAUUCGCCUGCUGUCAAUUUCUUGACACCCGGUAUUUCGCAUUUGCGACAUACAACAAAUACCGUGUGCAAUCGCUCAUAAAUCGGGUGUAGGCAA ...-------................((((((.(((((....)))))(((((((((.((((....)))).))))..........(((.((....)).)))....))))))))))).. ( -36.00) >DroMoj_CAF1 80995 107 + 1 CAUUUCGUUUAUUGCGGUAGA-GUUUGCCUGCUCGCCUUUUCUCAGCACCCAGCAGUUGGCAUACGCGCCGCACAACAAAAU---------UGUGCUCAUAAACCGUGUGUAGGCAA ....((((.....))))....-..((((((((.(((.........((.....)).((((((......)))((((((.....)---------))))).....))).))).)))))))) ( -30.00) >consensus AAU_______UUUUCGGUAGAUAUUCGCCUGCUGUCGAUUUCUUGACACCCGGUAUUUCGCAUUUGCGCCAUACAACAAAUACCAUG_G_AUUUGCUCAUAAAUCGGGUGUAGGCAA ..........................((((((((((((....))))))((((((.....(((.(((((...................))))).)))......)))))).)))))).. (-13.02 = -16.86 + 3.84)


| Location | 3,586,989 – 3,587,099 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.88 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3586989 110 - 22224390 UUGCCUCCGCCCGAUUUAUGAGCAAAUACCCAUGGUAUUUGUUGUAUGGAGCAAAUGCGAAAUACCGGGUGUCAAGAAAUCGACAGCAGGCGAAUAUCUACCAAAAA-------AUU ((((((.((((((.......((((((((((...))))))))))((((...((....))...)))))))))(((.(....).))).).))))))..............-------... ( -33.80) >DroVir_CAF1 76802 100 - 1 UUACCUACACACGGUUUAUGAGCACA---------AUUUUGUUGUAUGGCGCGUAUGCCAACUGCUGGGUGCUGAGAAAAGGCGAGCAGGCAAAC---UGCCGCAACA-----AAUG .((((((((...(((.((((.(((((---------((...)))))...)).)))).)))...)).))))))..........(((.((((.....)---))))))....-----.... ( -27.40) >DroSec_CAF1 45965 110 - 1 UUGCCUACGCCCGAUUUAUGAGCAAAUACCUAUGGUAUUUGUUGUAUGGAGCAAAUGCGAAAUACCGGGUGUCAAGAAAUCGACAGCAGGCGAAUAUCUACCAAAAA-------AUU ((((((.((((((.......((((((((((...))))))))))((((...((....))...)))))))))(((.(....).))).).))))))..............-------... ( -33.80) >DroEre_CAF1 40776 110 - 1 UUGCCUACACCCGAUUUAUGAGCGAUUGCACAUGGUAUUUGUUGUAUGUCGCAAAUGCGAAAUACCGGGUGUCAAGAAAUCGACAGCAGGCGAAUAUCUACCGAAAA-------AUU ((((((.((((((.......(((((.(((.....))).)))))((((.((((....)))).)))))))))(((.(....).))).).))))))..............-------... ( -31.80) >DroYak_CAF1 41684 110 - 1 UUGCCUACACCCGAUUUAUGAGCGAUUGCACACGGUAUUUGUUGUAUGUCGCAAAUGCGAAAUACCGGGUGUCAAGAAAUUGACAGCAGGCGAAUAUCUACCGAAAA-------AUU ((((((.((((((.......(((((.(((.....))).)))))((((.((((....)))).)))))))))(((((....))))).).))))))..............-------... ( -35.90) >DroMoj_CAF1 80995 107 - 1 UUGCCUACACACGGUUUAUGAGCACA---------AUUUUGUUGUGCGGCGCGUAUGCCAACUGCUGGGUGCUGAGAAAAGGCGAGCAGGCAAAC-UCUACCGCAAUAAACGAAAUG ..(((.......)))...........---------((((((((.(((((.(.((.((((...((((...((((.......)))))))))))).))-.)..)))))...)))))))). ( -27.90) >consensus UUGCCUACACCCGAUUUAUGAGCAAAU_C_CAUGGUAUUUGUUGUAUGGCGCAAAUGCGAAAUACCGGGUGUCAAGAAAUCGACAGCAGGCGAAUAUCUACCGAAAA_______AUU ((((((.((((((.......(((((((((.....)))))))))((((...((....))...))))))))))..........(....)))))))........................ (-20.06 = -20.95 + 0.89)

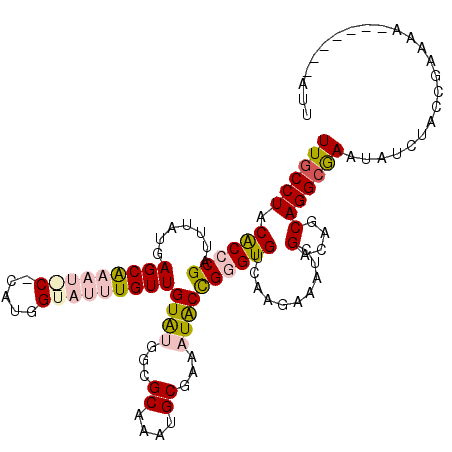
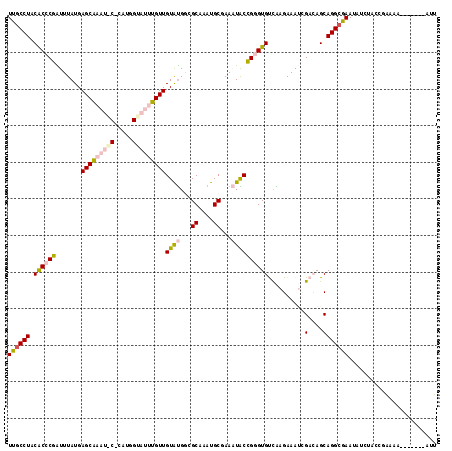
| Location | 3,587,020 – 3,587,130 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.45 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3587020 110 - 22224390 AUUAAUCUCCAGCACGAAAAUCUGCUAAUAUUUGCCUCCGCCCGAUUUAUGAGCAAAUACCCAUGGUAUUUGUUGUAUGGAGCAAAUGCGAAAUACCGGGUGUCAAGAAA .....(((..((((.(.....)))))............((((((.......((((((((((...))))))))))((((...((....))...))))))))))...))).. ( -28.20) >DroSec_CAF1 45996 110 - 1 AUUAAUCUCCAGCGCGAAAAUCUGCUAAUAUUUGCCUACGCCCGAUUUAUGAGCAAAUACCUAUGGUAUUUGUUGUAUGGAGCAAAUGCGAAAUACCGGGUGUCAAGAAA .....(((..((.(((((.((......)).)))))))(((((((.......((((((((((...))))))))))((((...((....))...)))))))))))..))).. ( -29.70) >DroSim_CAF1 32775 110 - 1 AUUAAUCUCCAGCACGAAAAUCUGCUAAUAUUUGCCUACGCCCGAUUUAUGAGCAAAUACCCAUGGUAUUUGUUGUAUGGAACAAAUGCGAAAUACCGGGUGUCAAGAAA ..........((((.(.....)))))....((((...(((((((.......((((((((((...))))))))))((((.......)))).......)))))))))))... ( -26.90) >DroEre_CAF1 40807 110 - 1 ACUAAUCCCUGGCACGAAAAUCAGCUAAUAUUUGCCUACACCCGAUUUAUGAGCGAUUGCACAUGGUAUUUGUUGUAUGUCGCAAAUGCGAAAUACCGGGUGUCAAGAAA ..........((((..................)))).(((((((.......(((((.(((.....))).)))))((((.((((....)))).)))))))))))....... ( -27.77) >DroYak_CAF1 41715 105 - 1 ACUAACU-----AAUGAAAAUCAGCUAAUAUUUGCCUACACCCGAUUUAUGAGCGAUUGCACACGGUAUUUGUUGUAUGUCGCAAAUGCGAAAUACCGGGUGUCAAGAAA .......-----........((.((........))..(((((((.......(((((.(((.....))).)))))((((.((((....)))).)))))))))))...)).. ( -25.80) >DroMoj_CAF1 81032 97 - 1 AUAAUAAAGCAGUGCGCA----ACACAAUAUUUGCCUACACACGGUUUAUGAGCACA---------AUUUUGUUGUGCGGCGCGUAUGCCAACUGCUGGGUGCUGAGAAA ...((..(((((((((((----(((.(((...((((.((.....))....).)))..---------))).))))))))((((....)))).))))))..))......... ( -26.90) >consensus AUUAAUCUCCAGCACGAAAAUCAGCUAAUAUUUGCCUACACCCGAUUUAUGAGCAAAUACCCAUGGUAUUUGUUGUAUGGAGCAAAUGCGAAAUACCGGGUGUCAAGAAA ..............................((((...(((((((.......(((((((((.....)))))))))((((...((....))...)))))))))))))))... (-19.07 = -19.45 + 0.38)

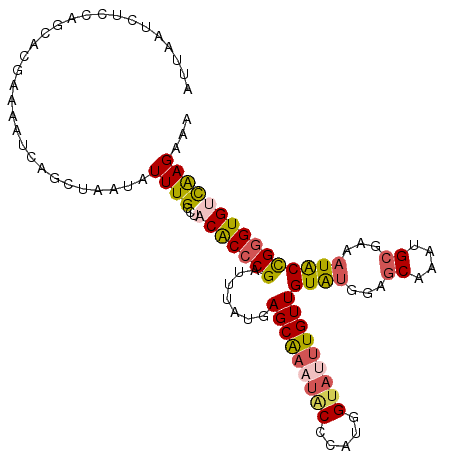
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:55 2006