
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,584,139 – 3,584,307 |
| Length | 168 |
| Max. P | 0.997249 |

| Location | 3,584,139 – 3,584,252 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.45 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -22.00 |
| Energy contribution | -23.04 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3584139 113 + 22224390 UAACCGCACUUGAAAUCACAUGAUUGACUGCUGUUCGAUUGCGGUUCGAUUAAUUCCGCUUGUGUGCAACAUGCA-------ACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAAC .(((((((.((((((((....)))).((....)))))).)))))))...........(((.(((((((...(((.-------...........)))...))).)))).)))......... ( -25.90) >DroSec_CAF1 43004 118 + 1 UAACCGGACUUGAAAUC--GUGAUUGACUGCUGUUCGAUUGCGGUUCGAUUACUUCCGCUUGUGUGCAACAUGCAACAUGCAACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAAC .....(((.........--((((((((((((.........)))).))))))))....(((.(((((((...(((....(((.....)))....)))...))).)))).)))..))).... ( -33.10) >DroSim_CAF1 29797 111 + 1 UAACCGCACUUGAAAUC--GUGAUUGACUGCUGUUCGAUUGCGGUUCGAUUACUUCCGCUUGUGUGCAACAUGCA-------ACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAAC .....((((........--((((((((((((.........)))).))))))))....(.(((((((...((((..-------.))))...)))))))))))).................. ( -28.90) >DroEre_CAF1 37765 111 + 1 UAACCCCACUUCAAAUC--GUGAUUGACUGCUGUUCGAUUGCAGUUCGAUUACUGCUGCUUGUGUGCAACAUGCA-------ACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAAC .................--((((((((((((.........)))).)))))))).((((..(((.((((...(((.-------...........)))...))))))).))))......... ( -27.50) >DroYak_CAF1 38800 111 + 1 AAACCCCACUUCAAAUC--GUGAUUGACUGCUGUUCGAUUGCGGCUCGAUUAACGCUGCUUGUGCGCAACAUGCA-------ACAUGCAACACGCAACGUGCAACAACAGCUUUCCAAAC .................--.(((((((..(((((......))))))))))))..((((.((((((((.....)).-------...(((.....)))....)).))))))))......... ( -25.60) >consensus UAACCGCACUUGAAAUC__GUGAUUGACUGCUGUUCGAUUGCGGUUCGAUUACUUCCGCUUGUGUGCAACAUGCA_______ACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAAC .....((((..........((((((((((((.........)))).))))))))....(.(((((((...((((..........))))...)))))))))))).................. (-22.00 = -23.04 + 1.04)

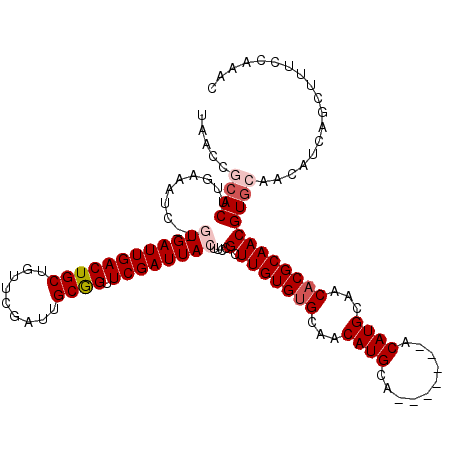

| Location | 3,584,179 – 3,584,292 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.44 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -25.53 |
| Energy contribution | -26.09 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3584179 113 + 22224390 GCGGUUCGAUUAAUUCCGCUUGUGUGCAACAUGCA-------ACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAACUGUGUCACAGAGUUGUUUUUGCCCGCAACUGACAGAACCA ..(((((((.....))((.(((((.((((...(((-------((.(((.....)))..(((..(((.(((.........)))))))))...)))))..)))).))))).))...))))). ( -29.60) >DroSec_CAF1 43042 120 + 1 GCGGUUCGAUUACUUCCGCUUGUGUGCAACAUGCAACAUGCAACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAACUGUGUCACAGAGUUGCUUUUGCCCGCAACUGACAGAACCA ..(((((..........(((.(((((((...(((....(((.....)))....)))...))).)))).)))...........(((((....(((((........))))))))))))))). ( -34.40) >DroSim_CAF1 29835 113 + 1 GCGGUUCGAUUACUUCCGCUUGUGUGCAACAUGCA-------ACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAACUGUGUCACAGAGUUGCUUUUGCCCGCAACUGACAGAACCA ..(((((((.....))((.(((((.((((...(((-------((.(((.....)))..(((..(((.(((.........)))))))))...)))))..)))).))))).))...))))). ( -31.20) >DroEre_CAF1 37803 113 + 1 GCAGUUCGAUUACUGCUGCUUGUGUGCAACAUGCA-------ACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAACUGUGUCACAGAGUUGCUCUUGCCCGCAACUGACAGAACCA (((((......)))))...(((((.((((...(((-------((.(((.....)))..(((..(((.(((.........)))))))))...)))))..)))).)))))............ ( -29.50) >DroYak_CAF1 38838 113 + 1 GCGGCUCGAUUAACGCUGCUUGUGCGCAACAUGCA-------ACAUGCAACACGCAACGUGCAACAACAGCUUUCCAAACUGUGUCACAGAGUUGUUUUUGCCCGCAACUGACAGAACCA ((((........(((.(((.(((..(((.......-------...))).))).))).)))((((.(((((((.......(((.....)))))))))).)))))))).............. ( -28.81) >consensus GCGGUUCGAUUACUUCCGCUUGUGUGCAACAUGCA_______ACAUGCAACACGCAACGUGCAACAUCAGCUUUCCAAACUGUGUCACAGAGUUGCUUUUGCCCGCAACUGACAGAACCA ..(((((..........(((.(((((((...(((...................)))...))).)))).)))...........(((((....(((((........))))))))))))))). (-25.53 = -26.09 + 0.56)


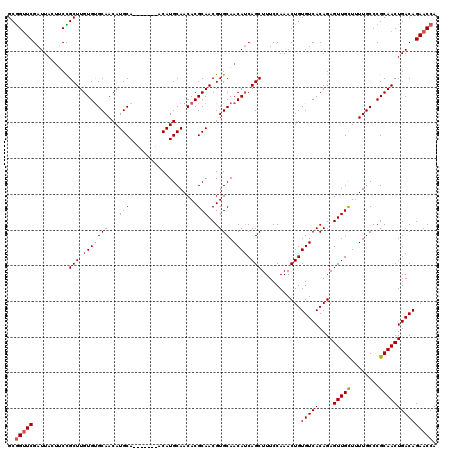
| Location | 3,584,179 – 3,584,292 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.44 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -37.56 |
| Energy contribution | -37.24 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3584179 113 - 22224390 UGGUUCUGUCAGUUGCGGGCAAAAACAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGU-------UGCAUGUUGCACACAAGCGGAAUUAAUCGAACCGC .(((((.((.((((.((........((((..((..((((((((.....))))).)))..)).)))).((((.(((((.-------..))))).)))).....)).)))).)).))))).. ( -35.60) >DroSec_CAF1 43042 120 - 1 UGGUUCUGUCAGUUGCGGGCAAAAGCAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGUUGCAUGUUGCAUGUUGCACACAAGCGGAAGUAAUCGAACCGC .(((((((((.......))))...(((((..((..((((((((.....))))).)))..)).)))))((((.((((((........)))))).))))....((....))....))))).. ( -42.30) >DroSim_CAF1 29835 113 - 1 UGGUUCUGUCAGUUGCGGGCAAAAGCAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGU-------UGCAUGUUGCACACAAGCGGAAGUAAUCGAACCGC .(((((((((.......))))...(((((..((..((((((((.....))))).)))..)).)))))((((.(((((.-------..))))).))))....((....))....))))).. ( -38.90) >DroEre_CAF1 37803 113 - 1 UGGUUCUGUCAGUUGCGGGCAAGAGCAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGU-------UGCAUGUUGCACACAAGCAGCAGUAAUCGAACUGC .........(((((.((.((((.((((((..((..((((((((.....))))).)))..)).))))).).))))..((-------(((.((((((......)))))))))))))))))). ( -39.70) >DroYak_CAF1 38838 113 - 1 UGGUUCUGUCAGUUGCGGGCAAAAACAACUCUGUGACACAGUUUGGAAAGCUGUUGUUGCACGUUGCGUGUUGCAUGU-------UGCAUGUUGCGCACAAGCAGCGUUAAUCGAGCCGC .(((((.....((.(((.((((...((((..((..((((((((.....))))).)))..)).))))....)))).)))-------.))(((((((......))))))).....))))).. ( -38.30) >consensus UGGUUCUGUCAGUUGCGGGCAAAAGCAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGU_______UGCAUGUUGCACACAAGCGGAAGUAAUCGAACCGC .(((((.((...((((........(((((..((..((((((((.....))))).)))..)).)))))((((.((((((........)))))).))))....)))).....)).))))).. (-37.56 = -37.24 + -0.32)


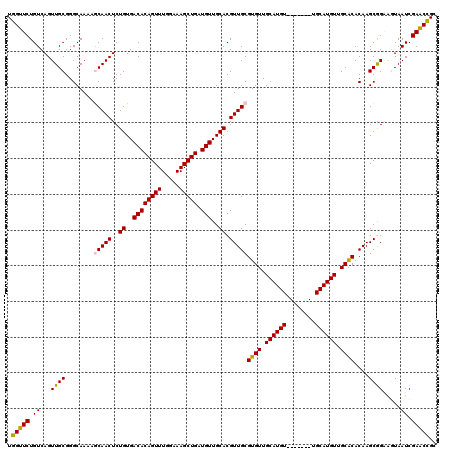
| Location | 3,584,214 – 3,584,307 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -31.86 |
| Energy contribution | -32.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3584214 93 - 22224390 -------------------------CGUUUGCCACCCAACUGGUUCUGUCAGUUGCGGGCAAAAACAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGU-- -------------------------..((((((.(.(((((((.....))))))).)))))))..((((..((..((((((((.....))))).)))..)).))))............-- ( -29.90) >DroSec_CAF1 43082 95 - 1 -------------------------CGUUUGCCACCCAACUGGUUCUGUCAGUUGCGGGCAAAAGCAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGUUG -------------------------..((((((.(.(((((((.....))))))).))))))).(((((..((..((((((((.....))))).)))..)).)))))............. ( -34.30) >DroSim_CAF1 29870 93 - 1 -------------------------CGUUUGCCACCCAACUGGUUCUGUCAGUUGCGGGCAAAAGCAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGU-- -------------------------..((((((.(.(((((((.....))))))).))))))).(((((..((..((((((((.....))))).)))..)).)))))...........-- ( -34.30) >DroEre_CAF1 37838 118 - 1 CCCACCAAUCUCCCUCCUUUGGCUUCGUUUUCCACCCAACUGGUUCUGUCAGUUGCGGGCAAGAGCAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGU-- ....((((..........))))............(((((((((.....))))))).))((((.((((((..((..((((((((.....))))).)))..)).))))).).))))....-- ( -35.50) >DroYak_CAF1 38873 115 - 1 ACCACCAAUCUCCC---CUCGCCAUCUUUUUCCACCCAACUGGUUCUGUCAGUUGCGGGCAAAAACAACUCUGUGACACAGUUUGGAAAGCUGUUGUUGCACGUUGCGUGUUGCAUGU-- ..............---.................(((((((((.....))))))).))((((...((((..((..((((((((.....))))).)))..)).))))....))))....-- ( -29.30) >consensus _________________________CGUUUGCCACCCAACUGGUUCUGUCAGUUGCGGGCAAAAGCAACUCUGUGACACAGUUUGGAAAGCUGAUGUUGCACGUUGCGUGUUGCAUGU__ ..................................(((((((((.....))))))).))((((..(((((..((..((((((((.....))))).)))..)).)))))...))))...... (-31.86 = -32.26 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:50 2006