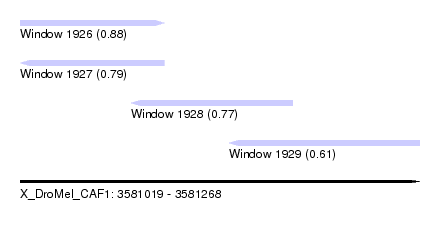
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,581,019 – 3,581,268 |
| Length | 249 |
| Max. P | 0.884881 |

| Location | 3,581,019 – 3,581,109 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -16.70 |
| Consensus MFE | -12.23 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
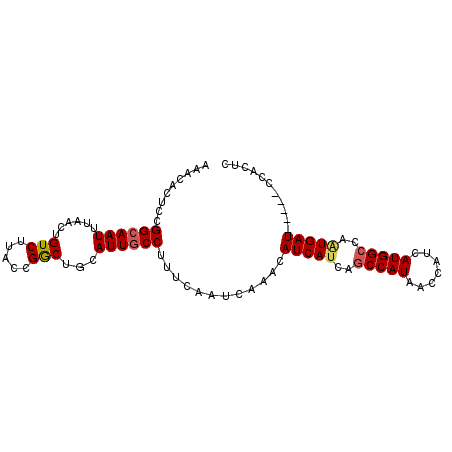
>X_DroMel_CAF1 3581019 90 + 22224390 AAACACUCCGGCAAUUUAACUGUCUUACCGGCUGCAUUACCUUUCAAUCAAACAUCAUCAGCCAUAACCAUCAUGGCCAUUGAU-----CCAUUC .......((((................))))......................((((...(((((.......)))))...))))-----...... ( -11.39) >DroSec_CAF1 39188 90 + 1 CUUCACUGUGGCAAUUUAACUGUCUUACCGACUGCAUUGCCUUUCAAUCAAACAUCAUCAGCCAUAACCAUCAUGGCCAGUGAU-----CCACUC ..((((((.((((((......(((.....)))...))))))...................(((((.......))))))))))).-----...... ( -21.30) >DroEre_CAF1 34295 95 + 1 AAACACUUGGGCAAUUUAACUGUCUUGCCGGCUGCAUUGCCCUUCAAUCAAACAUCAUCACCCAUAACCCACAUGGCCAAUGAUCCGAUCCGCUC ........(((((((.((.(((......))).)).)))))))...........(((((...((((.......))))...)))))........... ( -17.40) >consensus AAACACUCCGGCAAUUUAACUGUCUUACCGGCUGCAUUGCCUUUCAAUCAAACAUCAUCAGCCAUAACCAUCAUGGCCAAUGAU_____CCACUC .........((((((......(((.....)))...))))))............(((((..(((((.......)))))..)))))........... (-12.23 = -12.90 + 0.67)

| Location | 3,581,019 – 3,581,109 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -18.61 |
| Energy contribution | -18.50 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3581019 90 - 22224390 GAAUGG-----AUCAAUGGCCAUGAUGGUUAUGGCUGAUGAUGUUUGAUUGAAAGGUAAUGCAGCCGGUAAGACAGUUAAAUUGCCGGAGUGUUU ......-----((((.(((((((((...))))))))).))))((...(((....)))...))..(((((((..........)))))))....... ( -23.30) >DroSec_CAF1 39188 90 - 1 GAGUGG-----AUCACUGGCCAUGAUGGUUAUGGCUGAUGAUGUUUGAUUGAAAGGCAAUGCAGUCGGUAAGACAGUUAAAUUGCCACAGUGAAG ......-----.(((((((((((((...)))))))...................((((((((.(((.....))).))...)))))).)))))).. ( -27.00) >DroEre_CAF1 34295 95 - 1 GAGCGGAUCGGAUCAUUGGCCAUGUGGGUUAUGGGUGAUGAUGUUUGAUUGAAGGGCAAUGCAGCCGGCAAGACAGUUAAAUUGCCCAAGUGUUU ((((((((((((((((((.(((((.....))))).))))))..)))))))...(((((((..(((.(......).)))..)))))))...))))) ( -30.60) >consensus GAGUGG_____AUCAAUGGCCAUGAUGGUUAUGGCUGAUGAUGUUUGAUUGAAAGGCAAUGCAGCCGGUAAGACAGUUAAAUUGCCAAAGUGUUU ...........((((..(((((((.....)))))))..))))............((((((..(((.(......).)))..))))))......... (-18.61 = -18.50 + -0.11)



| Location | 3,581,088 – 3,581,189 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.45 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -26.67 |
| Energy contribution | -27.05 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3581088 101 - 22224390 UCGGCGUGAUUACGUGAGUAAUUCCCAAUGUUCGCCCAUGGGAAGACCAGCUUGAAAGGGUUAAGAUUGAGCCUAAGGUGGAAUGG-----AUCAAUGGCCAUGAU-------------- ..(((.(((((((....))..((((((.((......))))))))..(((.(((...(((.(((....))).)))))).)))....)-----))))...))).....-------------- ( -28.90) >DroSec_CAF1 39257 101 - 1 UCGGCUAAGUUACGUGAGUAAUUCCCAAUGUUCACCCAUGGCAAGACCAGCUUGAAAGGGUUAAGAUUGAGCCUAAGGUGGAGUGG-----AUCACUGGCCAUGAU-------------- ..(((((((((((....)))))).(((...((((((...(((..((.((((((....)))))..).))..)))...)))))).)))-----.....))))).....-------------- ( -31.90) >DroEre_CAF1 34364 106 - 1 UUGGCCAAGUUACGUGAGUAAUUCCCAAUGUUCACCCAAGGGAAGACCAGCUUGAUGGGGUUAAGAUUGAGCCUAGGGUGGAGCGGAUCGGAUCAUUGGCCAUGUG-------------- .((((((((((((....)))))(((...(((((((((..((.....)).......((((.(((....))).)))))))).)))))....)))...)))))))....-------------- ( -38.70) >DroYak_CAF1 35593 115 - 1 UUGGCCAAGAUACGUGAGUAAUUCCCAAUGUUCACCCAAGGGAAGACCAGCUUAAAAGGGUUAAGAUUGAGCCUAGGGUGGAGCUG-----AUCAUUGGCCAUGAGGCAUCAGGGCUAUG .(((((((..(((....))).(((((.............)))))((.((((((....(((((.......)))))......))))))-----.)).)))))))...(((......)))... ( -35.52) >consensus UCGGCCAAGUUACGUGAGUAAUUCCCAAUGUUCACCCAAGGGAAGACCAGCUUGAAAGGGUUAAGAUUGAGCCUAAGGUGGAGCGG_____AUCAUUGGCCAUGAG______________ ..(((((((((((....)))))).......(((((((..((.....))........(((.(((....))).))).)))))))..............)))))................... (-26.67 = -27.05 + 0.38)



| Location | 3,581,149 – 3,581,268 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -27.64 |
| Energy contribution | -29.39 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
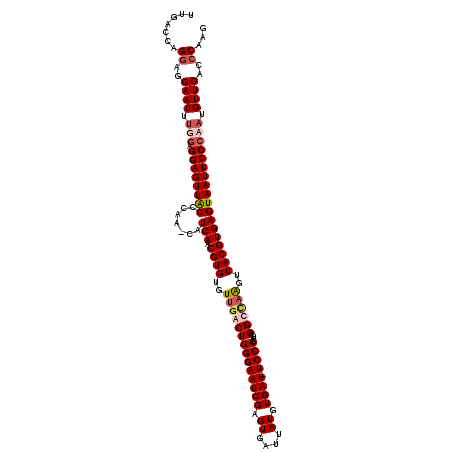
>X_DroMel_CAF1 3581149 119 - 22224390 UUGACCAGGAGGAAUUUGAAGGAGUUACCCAACCAUCAACGUGUGUUGACUGGGAAUCGAGUUAUUAUGUGAU-UCCCCCUCGGCGUGAUUACGUGAGUAAUUCCCAAUGUUCGCCCAUG .......((..((((.....((((((((.((...((((.....((((((..((((((((.((....)).))))-))))..))))))))))....)).))))))))....))))..))... ( -33.80) >DroSec_CAF1 39318 119 - 1 UUGACCAGGAGGAAUUCGGAGGAGUUACCCAACCAUCAACGUGUGUUGACUGGGAAUCGAGUUAUUAUGUGAU-UCCCCCUCGGCUAAGUUACGUGAGUAAUUCCCAAUGUUCACCCAUG .......((..((((...(.((((((((.......(((.((((.(((((..((((((((.((....)).))))-))))..))))).....))))))))))))))))...))))..))... ( -36.41) >DroEre_CAF1 34430 119 - 1 UUGACCAGGAGGAAUUUUGAGGAGUUACCCAA-CAUCAACGUGUGUUGGCUGGGAAUCGAGUGAUUAUGUGAUUUCCCCUUUGGCCAAGUUACGUGAGUAAUUCCCAAUGUUCACCCAAG .......((..((((.(((.((((((((....-..(((.((((..((((((((((((((.((....)).)))))))).....))))))..)))))))))))))))))).))))..))... ( -38.90) >DroYak_CAF1 35668 118 - 1 UUGACCAGAAGGAAUUUUGAGGAGUUGCCCAA-CAUCAACGUGUGUUGGCUGGGAAUCGAGUGAUUAUGUGAU-UCCCCUUUGGCCAAGAUACGUGAGUAAUUCCCAAUGUUCACCCAAG ...........((((.(((.((((((((....-..(((.(((((.((((((((((((((.((....)).))))-))))....)))))).))))))))))))))))))).))))....... ( -41.50) >consensus UUGACCAGGAGGAAUUUGGAGGAGUUACCCAA_CAUCAACGUGUGUUGACUGGGAAUCGAGUGAUUAUGUGAU_UCCCCCUCGGCCAAGUUACGUGAGUAAUUCCCAAUGUUCACCCAAG .......((..((((.(((.((((((((.......(((.((((..((((((((((((((.((....)).)))).))))....))))))..)))))))))))))))))).))))..))... (-27.64 = -29.39 + 1.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:45 2006