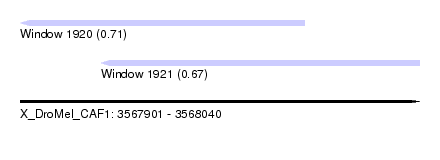
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,567,901 – 3,568,040 |
| Length | 139 |
| Max. P | 0.714523 |

| Location | 3,567,901 – 3,568,000 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 84.09 |
| Mean single sequence MFE | -19.99 |
| Consensus MFE | -14.65 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
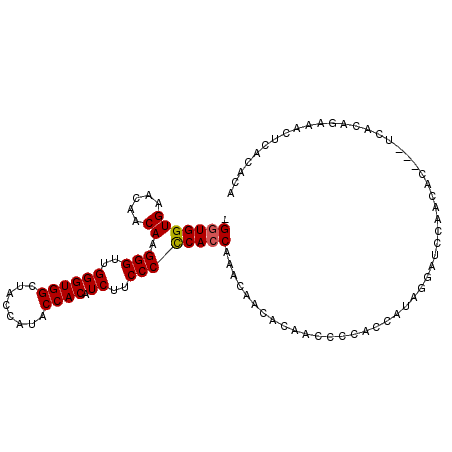
>X_DroMel_CAF1 3567901 99 - 22224390 GGGUGGUGAA---CAAGGGUUGGGUGGCUACCAUACCACAUCUUCCCCCAACAAACUCCACACCCCCCCCCC--CCCCCAACGCCCAUCACAGAAACACACACA (((.((((..---..((.((((((.((.................))))))))...))...))))))).....--.............................. ( -20.73) >DroSec_CAF1 24150 100 - 1 -GGUGGUGAACAACAAGGGUUGGGUGGCUACCAUACCACAUCUUCCCCCACCAAACAACACAACCCCAUCAUAGGAUCCAACAC---UCACAGAAACUCACACA -.((((((........(((((((((((....................)))))........)))))).(((....)))....)))---.)))............. ( -18.45) >DroSim_CAF1 21530 100 - 1 -GGUGGUGAACAACAAGGGUGGGGUGGCUACCAUACCACAUCUUCCCUCACCAAACAACACAACCCCUCCAUAGGAUCCAACAC---UCACAGAAACUCACACA -((((((((.....((((((((..(((...)))..)))).))))...))))))............(((....)))..)).....---................. ( -20.80) >consensus _GGUGGUGAACAACAAGGGUUGGGUGGCUACCAUACCACAUCUUCCCCCACCAAACAACACAACCCCACCAUAGGAUCCAACAC___UCACAGAAACUCACACA .(((((((.....)).(((..((((((........)))).))..)))))))).................................................... (-14.65 = -14.77 + 0.11)

| Location | 3,567,929 – 3,568,040 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.74 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -17.66 |
| Energy contribution | -20.18 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
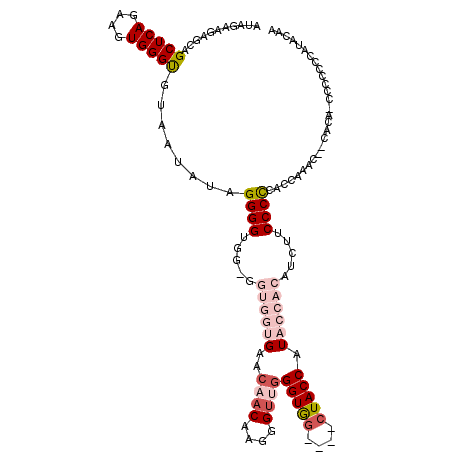
>X_DroMel_CAF1 3567929 111 - 22224390 AUAGAAGAGCAGCUCAGAAGUGGGUGUAAUAAAGGGGUGGGGGUGGUGAA---CAAGGGUUGGGUGG----CUACCAUACCACAUCUUCCCCCAACAAACUCCACACCCCCCCCCC--CC ........(((.((((....)))))))......((((.(((((.((((..---..((.((((((.((----.................))))))))...))...))))))))))))--). ( -40.73) >DroSec_CAF1 24175 115 - 1 AUAGAAGAGCAGCUCAGAAGUGGGUGUAAUAUAGGGGUGG-GGUGGUGAACAACAAGGGUUGGGUGG----CUACCAUACCACAUCUUCCCCCACCAAACAACACAACCCCAUCAUAGGA ......(((...))).((.(.((((..........(((((-((.((((..((((....))))((((.----......)))).))))...)))))))..........))))).))...... ( -33.95) >DroSim_CAF1 21555 115 - 1 AUAGAAGAGCAGCUCAGAAGUGGGUGUAAUAUAGGGGUGG-GGUGGUGAACAACAAGGGUGGGGUGG----CUACCAUACCACAUCUUCCCUCACCAAACAACACAACCCCUCCAUAGGA ........(((.((((....))))))).....((((((.(-..((((((.....((((((((..(((----...)))..)))).))))...))))))..)......))))))........ ( -33.70) >DroEre_CAF1 23948 93 - 1 AUUGAAAAGGAGCUCAGAAGUGGGUGGAAUAUGGGGGUGG-GGUGUUGAACAACAAGGGUUGGGUAG----CUACCAU---------UCCCCCAUC-------------CCCCCUUACAU .((((........))))(((.(((.(((...((((((...-(((((((..((((....))))..)))----).)))..---------.))))))))-------------))))))).... ( -36.40) >DroYak_CAF1 25521 101 - 1 AUUGAAGAGCAGCUCAGAAGUGGGCGGAAUAUAGGGGUGU-GGUGUUGAACAACAAGGGUUGGGUGGUUGGGUACCAU---------UCCCCCAUCUCU---------CCCCACUUACAU ......(((...)))..(((((((.(((.....((((.((-((((((.(((.((.........)).))).))))))))---------.))))....)))---------.))))))).... ( -32.60) >consensus AUAGAAGAGCAGCUCAGAAGUGGGUGUAAUAUAGGGGUGG_GGUGGUGAACAACAAGGGUUGGGUGG____CUACCAUACCACAUCUUCCCCCACCAAAC__CACA_CCCCCCCAUACAA ...........(((((....)))))........((((.....((((((..((((....))))(((((....))))).)))))).....))))............................ (-17.66 = -20.18 + 2.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:38 2006