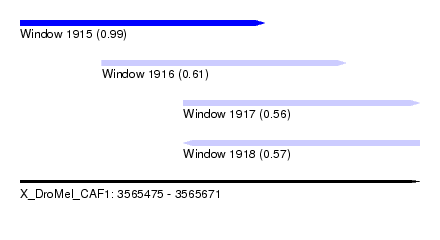
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,565,475 – 3,565,671 |
| Length | 196 |
| Max. P | 0.994765 |

| Location | 3,565,475 – 3,565,595 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -50.80 |
| Consensus MFE | -38.14 |
| Energy contribution | -37.58 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3565475 120 + 22224390 AUAUUGAGGUGGGUGUGAUCGCUAAGGCUGGUGGGUGUACUCGCUUUGCUGGUUGAUGUACUCGAUAGGCGAGCGAGCGAGUUGUGCACUCGCUGGCGAGCGGUUUAAGUAUCCGAUAAC .(((((.((((..((.(((((((...((..((((((((((((((((((((.(((((.....))))).))))...))))))))...))))))))..)).)))))))))..))))))))).. ( -51.60) >DroSec_CAF1 21767 120 + 1 AUAUUGAGGUGGGUGUGAUCGCUAAGGCUGGUGGGUGUACUCGCCAAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGAGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGC .......(((((.(..(((((((...((..((((((((((.((((.....))))(((...((((.........))))...)))))))))))))..)).)))))))..).)))))...... ( -48.90) >DroSim_CAF1 18554 120 + 1 AUAUUGAGGUGGGUGUGAUCGCUAAGGCUGGUGGGUGUACUCGCCAAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGCGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGC .......(((((.(..(((((((...((..((((((((((((((...(((.((..((.......))..)).)))..))))))...))))))))..)).)))))))..).)))))...... ( -49.20) >DroEre_CAF1 21537 116 + 1 AUAGCGAGGUGGGUGUCGUCGCUAAGGUCGGUAGGUGUACUGGCUAAGCUGGUGCAUGUACUCGGUGGGCGAG----CGAUUCGUGCAUUCGCUGGUACGCGGUGUAUGUCCUCGCUAAA .((((((((.....(((((((((....((((((.((((((..((...))..)))))).))).)))..))))).----)))).(((((((.(((......)))))))))).)))))))).. ( -53.50) >consensus AUAUUGAGGUGGGUGUGAUCGCUAAGGCUGGUGGGUGUACUCGCCAAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGCGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAAC .((((..(((((....(((((((...(((((((((((((((((((.....))))).....((((.....))))..........)))))))))))))).)))))))....))))))))).. (-38.14 = -37.58 + -0.56)



| Location | 3,565,515 – 3,565,635 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -44.80 |
| Consensus MFE | -27.81 |
| Energy contribution | -29.12 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3565515 120 + 22224390 UCGCUUUGCUGGUUGAUGUACUCGAUAGGCGAGCGAGCGAGUUGUGCACUCGCUGGCGAGCGGUUUAAGUAUCCGAUAACUAUGGUUGGUGCACACGCUAAGUAACAAGUGACCGCUAAA .....(((((.(((((.....))))).)))))((.(((((((.....))))))).)).((((((....(((.(((((.......))))))))...((((........))))))))))... ( -41.40) >DroSec_CAF1 21807 120 + 1 UCGCCAAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGAGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGCUAUGGGUGGUGCACUCGCUAAGUAACAAGUGACCGCCAAA ((((((.((.((((.(((.((((....(.....)....))))))).)))).))))))))((((((...(((((...((((...(((((...))))))))).)))))....)))))).... ( -46.70) >DroSim_CAF1 18594 120 + 1 UCGCCAAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGCGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGCUAUAGGUGGUGCACUCGCUAAGUAACAAGUGACCGCCAAA ((((((.((.((((.(((.(((((...(.....)...)))))))).)))).))))))))((((((...(((((...((((....((((...)))).)))).)))))....)))))).... ( -47.10) >DroEre_CAF1 21577 116 + 1 UGGCUAAGCUGGUGCAUGUACUCGGUGGGCGAG----CGAUUCGUGCAUUCGCUGGUACGCGGUGUAUGUCCUCGCUAAAUGUGGUUUGUGCGCUCGCUGCAUUGCAUGUGAUUGCCAAA ((((..(((.((((((((.(((((...((((((----.((..(((((((.(((......))))))))))))))))))...)).))).)))))))).))).(((.....)))...)))).. ( -44.00) >consensus UCGCCAAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGCGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAACUAUGGGUGGUGCACUCGCUAAGUAACAAGUGACCGCCAAA ((((((.((.((((.(((.(((((...(.....)...)))))))).)))).))))))))((((((...(((((.....((((....))))((....))...)))))....)))))).... (-27.81 = -29.12 + 1.31)


| Location | 3,565,555 – 3,565,671 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.43 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -16.00 |
| Energy contribution | -19.32 |
| Covariance contribution | 3.32 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3565555 116 + 22224390 GUUGUGCACUCGCUGGCGAGCGGUUUAAGUAUCCGAUAACUAUGGUUGGUGCACACGCUAAGUAACAAGUGACCGCUAAACUGCUACUUUAACAGACACAACUGUGCAUGUUAAGC .....((....((.(...((((((....(((.(((((.......))))))))...((((........))))))))))...).))....((((((..(((....)))..)))))))) ( -29.70) >DroSec_CAF1 21847 116 + 1 GUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGCUAUGGGUGGUGCACUCGCUAAGUAACAAGUGACCGCCAAACCGCCACUUUAACAGACACAACUGUGCAUGUUAAGC ((((.((....))))))(.(((((((.(((((....)))))...((((((.((((.(........).))))))))))))))))))..(((((((..(((....)))..))))))). ( -39.90) >DroSim_CAF1 18634 116 + 1 GUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGCUAUAGGUGGUGCACUCGCUAAGUAACAAGUGACCGCCAAACCGCUACUUUAACAGGCACAACUGUGCAUGUUAAGC ((((.((....)))))).((((((((.(((((....)))))...((((((.((((.(........).))))))))))))))))))..(((((((.((((....)))).))))))). ( -44.20) >DroEre_CAF1 21613 114 + 1 UUCGUGCAUUCGCUGGUACGCGGUGUAUGUCCUCGCUAAAUGUGGUUUGUGCGCUCGCUGCAUUGCAUGUGAUUGCCAAACCGCGCC--AAACAGUCACAAUUGUGCAUGUUAAGC ..(((((((.(((......)))))))))).....(((....((((((((.(((.(((((((...))).)))).)))))))))))...--.((((..(((....)))..)))).))) ( -38.20) >DroYak_CAF1 23252 95 + 1 ---------------------GGGGCAAGUCCCUAGUAGCUAUGGUUUGUGUACUCGCUGAGUAACAGGUGACUGCCAAACCGCUACUUUAACAGUCACAAUGGUGCAUGUUAAGC ---------------------((((....)))).((((((...((((((.(((.(((((........))))).)))))))))))))))((((((..(((....)))..)))))).. ( -33.50) >consensus GUCGUGCACUCGCUGGCGAGCGGUUUAAGUCACCGGUAGCUAUGGUUGGUGCACUCGCUAAGUAACAAGUGACCGCCAAACCGCUACUUUAACAGACACAACUGUGCAUGUUAAGC ...(((((((...((((...(((((.....)))))...)))).....)))))))(((((........)))))..((......))....((((((..(((....)))..)))))).. (-16.00 = -19.32 + 3.32)


| Location | 3,565,555 – 3,565,671 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.43 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -16.38 |
| Energy contribution | -19.96 |
| Covariance contribution | 3.58 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3565555 116 - 22224390 GCUUAACAUGCACAGUUGUGUCUGUUAAAGUAGCAGUUUAGCGGUCACUUGUUACUUAGCGUGUGCACCAACCAUAGUUAUCGGAUACUUAAACCGCUCGCCAGCGAGUGCACAAC ((((((((.((((....)))).)))))(((((((((............)))))))))))).(((((((...((.........))..........((((....)))).))))))).. ( -31.90) >DroSec_CAF1 21847 116 - 1 GCUUAACAUGCACAGUUGUGUCUGUUAAAGUGGCGGUUUGGCGGUCACUUGUUACUUAGCGAGUGCACCACCCAUAGCUACCGGUAACUUAAACCGCUCGCCAGCGAGUGCACGAC ((((((((.((((....)))).)))))).(((((.....((.(((((((((((....)))))))).))).))....))))).(((.......)))(((((....)))))))..... ( -41.60) >DroSim_CAF1 18634 116 - 1 GCUUAACAUGCACAGUUGUGCCUGUUAAAGUAGCGGUUUGGCGGUCACUUGUUACUUAGCGAGUGCACCACCUAUAGCUACCGGUAACUUAAACCGCUCGCCAGCGAGUGCACGAC ((((((((.((((....)))).)))))).(((((.....((.(((((((((((....)))))))).))).))....))))).(((.......)))(((((....)))))))..... ( -44.10) >DroEre_CAF1 21613 114 - 1 GCUUAACAUGCACAAUUGUGACUGUUU--GGCGCGGUUUGGCAAUCACAUGCAAUGCAGCGAGCGCACAAACCACAUUUAGCGAGGACAUACACCGCGUACCAGCGAAUGCACGAA (((.((((..(((....)))..)))).--)))((((((((((..((.(.(((...)))).))..)).)))))).......(((..(.....)..)))......))........... ( -30.80) >DroYak_CAF1 23252 95 - 1 GCUUAACAUGCACCAUUGUGACUGUUAAAGUAGCGGUUUGGCAGUCACCUGUUACUCAGCGAGUACACAAACCAUAGCUACUAGGGACUUGCCCC--------------------- ..((((((..(((....)))..))))))((((((((((((((((....))))(((((...)))))..))))))...)))))).(((.....))).--------------------- ( -28.80) >consensus GCUUAACAUGCACAGUUGUGACUGUUAAAGUAGCGGUUUGGCGGUCACUUGUUACUUAGCGAGUGCACCAACCAUAGCUACCGGGAACUUAAACCGCUCGCCAGCGAGUGCACGAC ((((((((.((((....)))).)))))(((((((((............))))))))))))..((((((..............((((.....))))((......))..))))))... (-16.38 = -19.96 + 3.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:35 2006