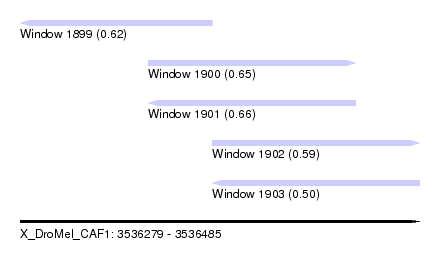
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,536,279 – 3,536,485 |
| Length | 206 |
| Max. P | 0.655249 |

| Location | 3,536,279 – 3,536,378 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.28 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
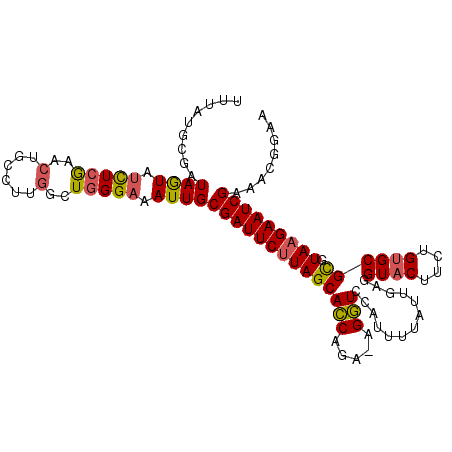
>X_DroMel_CAF1 3536279 99 - 22224390 UGUGCGGAAAUCAAAGGGAAACCUU--UGUGGGACAAAACCGGAACAGGACUCGAA-UGAGGGGUUGGUU-GUUGCGCACACAAGAGGUGAUUGAGAUUCCCC (((((......((((((....))))--)).(.((((..(((((..(....(((...-.))))..))))))-))).)))))).....((.((.......)))). ( -27.80) >DroSec_CAF1 44449 95 - 1 UGUGCGGAAAUCAAAGGGAAACCUU--UCUGGGACAAAACCGGAACAGGACUCGAA-AGAGGGGU----U-GUUGCACACACAAGAGGUGAUUGAGAUUCCCC (((((((.((((.((((....))))--(((((.......)))))......(((...-.))).)))----)-.)))))))(((.....)))............. ( -30.30) >DroSim_CAF1 45643 95 - 1 UGUGCGGAAAUCAAAGGGAAACCUU--UGUGGGACAAAACCGGAACAGGACUCGAA-AGAGGGGU----U-GUUGCACACACAAGAGGUGAUUGAGAUUCCCC .....((.(((((((((....))))--)(((.(((((..((......)).(((...-.)))...)----)-))).))).(((.....))).....)))).)). ( -30.80) >DroEre_CAF1 48743 99 - 1 UGUACGGAAAUCAAAGGGAAACCUCUCUGCGGGACAAAACCGGAACAGGACUCGAAAAGAGGGGU----UGGAUGCACACACGAGAGGUGAUUGAGAUUCCCC .....((((.((((.....(((((((((.((((......((......)).))))...))))))))----).......(((.(....)))).))))..)))).. ( -27.90) >DroYak_CAF1 44482 95 - 1 UGUGCGGAAAUCAAAGGGAAACCUU--UGUGGGACAAAACCGGAACAGGACUCGAA-AGAGGGGU----U-GAUGCACACACGAGAGGUGAUUGAGAUUCCCC ((((((..(((((((((....))))--)...........((......)).(((...-.))).)))----)-..))))))(((.....)))............. ( -28.20) >consensus UGUGCGGAAAUCAAAGGGAAACCUU__UGUGGGACAAAACCGGAACAGGACUCGAA_AGAGGGGU____U_GUUGCACACACAAGAGGUGAUUGAGAUUCCCC .....((((.(((((((....)))...((((.(((....((......)).(((.....)))..........))).))))(((.....))).))))..)))).. (-21.60 = -22.28 + 0.68)



| Location | 3,536,345 – 3,536,452 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -19.85 |
| Energy contribution | -21.60 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3536345 107 + 22224390 GUCCCACAAAGGUUUCCCUUUGAUUUCCGCACAUUCCGUUUCGAUUCUUACGCGCACAGAAGUACCUCAAUAAAGUGGACCA-ACUGGUGCUAAGAAUCGCAAUUUCC ......((((((....))))))...............(((.(((((((((.((((.(((..(..((.(......).))..).-.)))))))))))))))).))).... ( -28.60) >DroSec_CAF1 44511 107 + 1 GUCCCAGAAAGGUUUCCCUUUGAUUUCCGCACAUUCCGUUUCGAUUCAUAAACGCACAGAAGUACCUCAAUAAAAUGGACCU-UCUGGUGCUAAGAAUCGCAAUUUCC .......(((((....)))))(((((..((((....(((((........)))))..((((((..((..........))..))-))))))))...)))))......... ( -23.30) >DroSim_CAF1 45705 107 + 1 GUCCCACAAAGGUUUCCCUUUGAUUUCCGCACAUUCCGUUUCGAUUCUUACGCGCACAGAAGUACCUCAAUAAAAUGGACCU-UCCGGUGCUAAGAAUCGCAAUUUCC ......((((((....))))))...............(((.(((((((((.((((.(.((((..((..........))..))-)).)))))))))))))).))).... ( -29.00) >DroYak_CAF1 44544 108 + 1 GUCCCACAAAGGUUUCCCUUUGAUUUCCGCACAUUCCGUUUCGAUUCUUACGCGCACAGCAGAACCUCCAUAAAAUGGAUCUUUCUGAUGCUAAGAAUCGCAAUAUCC ......((((((....))))))...............(((.(((((((((.(((.....(((((..(((((...)))))...))))).)))))))))))).))).... ( -27.50) >consensus GUCCCACAAAGGUUUCCCUUUGAUUUCCGCACAUUCCGUUUCGAUUCUUACGCGCACAGAAGUACCUCAAUAAAAUGGACCU_UCUGGUGCUAAGAAUCGCAAUUUCC ......((((((....))))))...............(((.(((((((((.((((.((((....((..........)).....))))))))))))))))).))).... (-19.85 = -21.60 + 1.75)

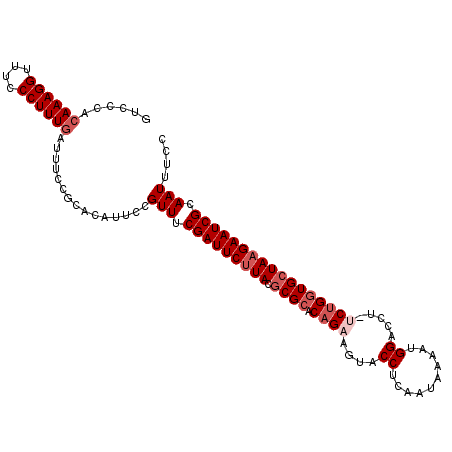
| Location | 3,536,345 – 3,536,452 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.57 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3536345 107 - 22224390 GGAAAUUGCGAUUCUUAGCACCAGU-UGGUCCACUUUAUUGAGGUACUUCUGUGCGCGUAAGAAUCGAAACGGAAUGUGCGGAAAUCAAAGGGAAACCUUUGUGGGAC ......(((........)))(((..-...(((.(((....)))((((((((((...((.......))..)))))).)))))))...((((((....)))))))))... ( -31.10) >DroSec_CAF1 44511 107 - 1 GGAAAUUGCGAUUCUUAGCACCAGA-AGGUCCAUUUUAUUGAGGUACUUCUGUGCGUUUAUGAAUCGAAACGGAAUGUGCGGAAAUCAAAGGGAAACCUUUCUGGGAC ........((((((...((((.(((-((..((..........))..)))))))))......))))))............(((((.....(((....)))))))).... ( -28.40) >DroSim_CAF1 45705 107 - 1 GGAAAUUGCGAUUCUUAGCACCGGA-AGGUCCAUUUUAUUGAGGUACUUCUGUGCGCGUAAGAAUCGAAACGGAAUGUGCGGAAAUCAAAGGGAAACCUUUGUGGGAC ........(((((((((((.(((((-((..((..........))..)))))).).)).)))))))))...................((((((....))))))...... ( -33.10) >DroYak_CAF1 44544 108 - 1 GGAUAUUGCGAUUCUUAGCAUCAGAAAGAUCCAUUUUAUGGAGGUUCUGCUGUGCGCGUAAGAAUCGAAACGGAAUGUGCGGAAAUCAAAGGGAAACCUUUGUGGGAC (.(((((.((((((((((((((((((...(((((...)))))..)))))..))))...)))))))))......))))).)......((((((....))))))...... ( -34.70) >consensus GGAAAUUGCGAUUCUUAGCACCAGA_AGGUCCAUUUUAUUGAGGUACUUCUGUGCGCGUAAGAAUCGAAACGGAAUGUGCGGAAAUCAAAGGGAAACCUUUGUGGGAC ........((((((((((((((.....))).............((((....)))))).)))))))))...................((((((....))))))...... (-25.39 = -25.57 + 0.19)



| Location | 3,536,378 – 3,536,485 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -14.85 |
| Energy contribution | -16.35 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3536378 107 + 22224390 UUCCGUUUCGAUUCUUACGCGCACAGAAGUACCUCAAUAAAGUGGACCA-ACUGGUGCUAAGAAUCGCAAUUUCCCAGCCAAAACAGUAUGAGAUACUAUCGCAUAAA ....(((.(((((((((.((((.(((..(..((.(......).))..).-.)))))))))))))))).)))................((((.(((...))).)))).. ( -24.70) >DroSec_CAF1 44544 107 + 1 UUCCGUUUCGAUUCAUAAACGCACAGAAGUACCUCAAUAAAAUGGACCU-UCUGGUGCUAAGAAUCGCAAUUUCCCAGCCAAGGCAGUUCGAGAUACUAUCGCAUAAA ....((((((((((.((..(((.((((((..((..........))..))-))))))).)).)))))...........((....)).....)))))............. ( -23.10) >DroSim_CAF1 45738 107 + 1 UUCCGUUUCGAUUCUUACGCGCACAGAAGUACCUCAAUAAAAUGGACCU-UCCGGUGCUAAGAAUCGCAAUUUCCCAGCCAAGGCAGUUCGAGAUACUAUCGCAUAAA ....(((((((((((((.((((.(.((((..((..........))..))-)).)))))))))))))...........((....)).....)))))............. ( -27.40) >DroYak_CAF1 44577 97 + 1 UUCCGUUUCGAUUCUUACGCGCACAGCAGAACCUCCAUAAAAUGGAUCUUUCUGAUGCUAAGAAUCGCAAUAUCCG--UGAAAGCA-----AAUUAUUAUUGUA---- ....(((.(((((((((.(((.....(((((..(((((...)))))...))))).)))))))))))).)))....(--(....)).-----.............---- ( -22.90) >consensus UUCCGUUUCGAUUCUUACGCGCACAGAAGUACCUCAAUAAAAUGGACCU_UCUGGUGCUAAGAAUCGCAAUUUCCCAGCCAAAGCAGUUCGAGAUACUAUCGCAUAAA ....(((.(((((((((.((((.((((....((..........)).....))))))))))))))))).)))..................................... (-14.85 = -16.35 + 1.50)


| Location | 3,536,378 – 3,536,485 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.95 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -18.85 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3536378 107 - 22224390 UUUAUGCGAUAGUAUCUCAUACUGUUUUGGCUGGGAAAUUGCGAUUCUUAGCACCAGU-UGGUCCACUUUAUUGAGGUACUUCUGUGCGCGUAAGAAUCGAAACGGAA ((((((((..(((((((((...((..(..(((((.....(((........))))))))-..)..))......)))))))))......))))))))............. ( -31.80) >DroSec_CAF1 44544 107 - 1 UUUAUGCGAUAGUAUCUCGAACUGCCUUGGCUGGGAAAUUGCGAUUCUUAGCACCAGA-AGGUCCAUUUUAUUGAGGUACUUCUGUGCGUUUAUGAAUCGAAACGGAA .....(((..((((((((((.........((((((((.......))))))))(((...-.)))........))))))))))..))).(((((........)))))... ( -26.90) >DroSim_CAF1 45738 107 - 1 UUUAUGCGAUAGUAUCUCGAACUGCCUUGGCUGGGAAAUUGCGAUUCUUAGCACCGGA-AGGUCCAUUUUAUUGAGGUACUUCUGUGCGCGUAAGAAUCGAAACGGAA ((((((((..((((((((((.........((((((((.......))))))))(((...-.)))........))))))))))......))))))))............. ( -33.70) >DroYak_CAF1 44577 97 - 1 ----UACAAUAAUAAUU-----UGCUUUCA--CGGAUAUUGCGAUUCUUAGCAUCAGAAAGAUCCAUUUUAUGGAGGUUCUGCUGUGCGCGUAAGAAUCGAAACGGAA ----.............-----....(((.--((.......((((((((((((((((((...(((((...)))))..)))))..))))...)))))))))...))))) ( -24.70) >consensus UUUAUGCGAUAGUAUCUCGAACUGCCUUGGCUGGGAAAUUGCGAUUCUUAGCACCAGA_AGGUCCAUUUUAUUGAGGUACUUCUGUGCGCGUAAGAAUCGAAACGGAA .........((((.(((((..(......)..))))).))))((((((((((((((.....))).............((((....)))))).)))))))))........ (-18.85 = -19.10 + 0.25)

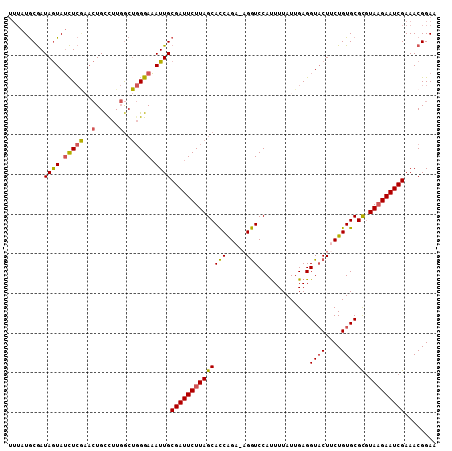
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:21 2006