
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,521,214 – 3,521,459 |
| Length | 245 |
| Max. P | 0.925304 |

| Location | 3,521,214 – 3,521,319 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -14.94 |
| Energy contribution | -16.28 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3521214 105 + 22224390 UCCGAUCAGUAAGUAAAUAAGUUUUGAACUAGGUUUGGGUAG----------UUGAUUUAAUUAGUCACCUUAAACAGUUCAAACAGUAUGAAAGCCUAGUAUAUACAUUUAUAU ............((((((..((((.(((((..((((((((..----------(((((...)))))..))).)))))))))))))).(((((...........))))))))))).. ( -17.50) >DroSec_CAF1 29572 102 + 1 UCCGAUCAGUAAGUACAUACAUUUUGAACUGGGGUUGGGUAGCUUAUCAUAUUUGAUUUAAUUAGUUACCUUAAACAGUUCAAACAGU-------------GCAUACAUAUAACU ........(((.((((......(((((((((.....((((((((.((((....))))......))))))))....)))))))))..))-------------)).)))........ ( -27.30) >DroSim_CAF1 30800 102 + 1 UCCGAUCAGUAAGUACAUACAUUUUGAACUGGGGUUGGGUAGCUUAUCAUAUUUGAUUUAAUUAGUUACCUUAAACAGUUCAAACAGU-------------GCAUACAUUUAACU ........(((.((((......(((((((((.....((((((((.((((....))))......))))))))....)))))))))..))-------------)).)))........ ( -27.30) >consensus UCCGAUCAGUAAGUACAUACAUUUUGAACUGGGGUUGGGUAGCUUAUCAUAUUUGAUUUAAUUAGUUACCUUAAACAGUUCAAACAGU_____________GCAUACAUUUAACU ......................(((((((((.....((((((((...................))))))))....)))))))))............................... (-14.94 = -16.28 + 1.33)

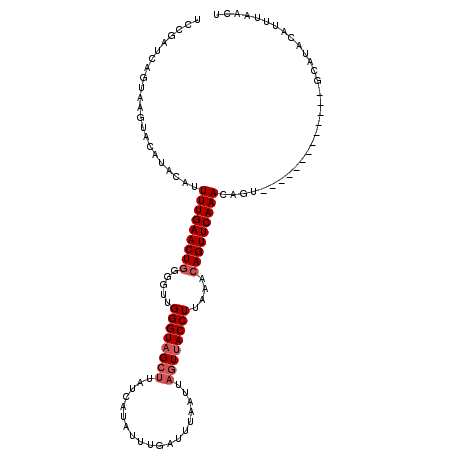
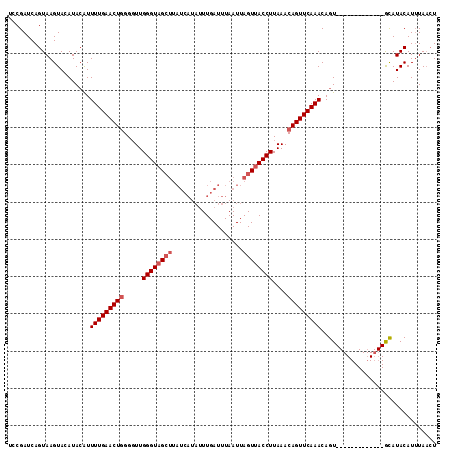
| Location | 3,521,279 – 3,521,398 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.53 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3521279 119 - 22224390 CGUCGCUGACGUCAGCGGCGUAAUAAAGCAAUA-AAUUGCAAUGUUAACUGAAGGGAUUAAGACGGAAACCAUUAAUGUAAUAUAAAUGUAUAUACUAGGCUUUCAUACUGUUUGAACUG ((((((((....))))))))(((((..((((..-..))))..)))))..((((((.(((((...(....)..)))))(((((((....)))).)))....)))))).............. ( -26.10) >DroSec_CAF1 29647 106 - 1 CGUCGCUGACGUCAGCGGCGUAACAGAGCAAUA-AAUUGCAAUGUUAACUCAAGGGAUUAAGACGGAAACCAUUAAUGUAAGUUAUAUGUAUGC-------------ACUGUUUGAACUG ((((((((....)))))))).(((((.(((...-...((((....(((((....(.(((((...(....)..))))).).)))))..)))))))-------------.)))))....... ( -27.60) >DroSim_CAF1 30875 106 - 1 CGUCGCUGACGUCAGCGGCGUAACAGAGCAAUA-AAUUGCAAUGUUAACUCAAGGGAUUAAGACGGAAACCAUUAAUGUAAGUUAAAUGUAUGC-------------ACUGUUUGAACUG ((((((((....)))))))).(((((.(((...-...((((...((((((....(.(((((...(....)..))))).).)))))).)))))))-------------.)))))....... ( -29.20) >DroYak_CAF1 28543 97 - 1 CGUCGCUGACGUCAGCGGCGUAACAAAGCAAAAAAAAUGCAAUGUUAACUGAAGGAAUUAAGACGGAAACCAUG--UGUAAAGG--------GG-------------AUUAUUACAAAUC ((((((((....))))))))(((((..(((.......)))..))))).................(....)....--(((((...--------..-------------....))))).... ( -20.20) >consensus CGUCGCUGACGUCAGCGGCGUAACAAAGCAAUA_AAUUGCAAUGUUAACUCAAGGGAUUAAGACGGAAACCAUUAAUGUAAGUUAAAUGUAUGC_____________ACUGUUUGAACUG ((((((((....))))))))(((((..((((.....))))..)))))......((..((......))..))................................................. (-20.46 = -20.53 + 0.06)

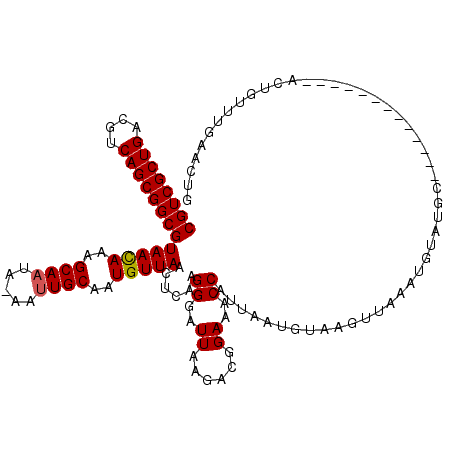
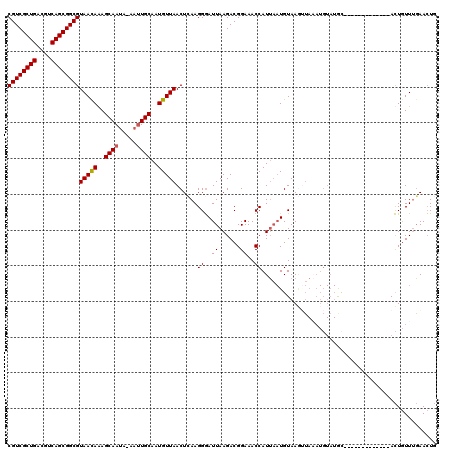
| Location | 3,521,319 – 3,521,420 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -26.46 |
| Energy contribution | -26.78 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.844981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3521319 101 - 22224390 CCUCUG------------------CUUGCAGCGCUGCCGGCGUCGCUGACGUCAGCGGCGUAAUAAAGCAAUA-AAUUGCAAUGUUAACUGAAGGGAUUAAGACGGAAACCAUUAAUGUA ((((..------------------...((((((((((.((((((...)))))).)))))))......((((..-..))))..))).....).))).(((((...(....)..)))))... ( -30.40) >DroSec_CAF1 29674 101 - 1 CCUCUG------------------CUUGCAGCGCUGCCGGCGUCGCUGACGUCAGCGGCGUAACAGAGCAAUA-AAUUGCAAUGUUAACUCAAGGGAUUAAGACGGAAACCAUUAAUGUA ((..((------------------......(((((((.((((((...)))))).)))))))((((..((((..-..))))..))))....))..))(((((...(....)..)))))... ( -32.40) >DroSim_CAF1 30902 101 - 1 CCUCUG------------------CUUGCAGCGCUGCCGGCGUCGCUGACGUCAGCGGCGUAACAGAGCAAUA-AAUUGCAAUGUUAACUCAAGGGAUUAAGACGGAAACCAUUAAUGUA ((..((------------------......(((((((.((((((...)))))).)))))))((((..((((..-..))))..))))....))..))(((((...(....)..)))))... ( -32.40) >DroYak_CAF1 28562 118 - 1 CCUCUGAUAACGUCGACUGCGACGCUUGCAGCACUGCCGGCGUCGCUGACGUCAGCGGCGUAACAAAGCAAAAAAAAUGCAAUGUUAACUGAAGGAAUUAAGACGGAAACCAUG--UGUA (((((((...(((((...((((((((.(((....))).)))))))))))))))))(((..(((((..(((.......)))..))))).))).))).........(....)....--.... ( -35.90) >consensus CCUCUG__________________CUUGCAGCGCUGCCGGCGUCGCUGACGUCAGCGGCGUAACAAAGCAAUA_AAUUGCAAUGUUAACUCAAGGGAUUAAGACGGAAACCAUUAAUGUA (((((.........................(((((((.((((((...)))))).)))))))((((..((((.....))))..))))..............))).)).............. (-26.46 = -26.78 + 0.31)


| Location | 3,521,359 – 3,521,459 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -23.45 |
| Energy contribution | -24.45 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3521359 100 - 22224390 GUACAAAACCG-CCCCAUAAAAUGAGACCCUGUUACAACGCCUCUG------------------CUUGCAGCGCUGCCGGCGUCGCUGACGUCAGCGGCGUAAUAAAGCAAUA-AAUUGC ...........-...........(((...............)))((------------------(((...(((((((.((((((...)))))).)))))))....)))))...-...... ( -26.26) >DroSec_CAF1 29714 101 - 1 UUCCUAGACCCCCCCCAUAAAAUGAGACUGUGUUACAACGCCUCUG------------------CUUGCAGCGCUGCCGGCGUCGCUGACGUCAGCGGCGUAACAGAGCAAUA-AAUUGC .........................(.((.((((((.........(------------------((.(((....))).)))(((((((....))))))))))))).)))....-...... ( -29.80) >DroSim_CAF1 30942 100 - 1 UUCCUAGACCC-CCCCAUAAAAUGAGACUCUGUUACAACGCCUCUG------------------CUUGCAGCGCUGCCGGCGUCGCUGACGUCAGCGGCGUAACAGAGCAAUA-AAUUGC ...........-.............(.(((((((((.........(------------------((.(((....))).)))(((((((....)))))))))))))))))....-...... ( -33.90) >DroYak_CAF1 28600 117 - 1 UUCCUGAGC---CCCCAUAAAGAGAGACUGUGUUACAACGCCUCUGAUAACGUCGACUGCGACGCUUGCAGCACUGCCGGCGUCGCUGACGUCAGCGGCGUAACAAAGCAAAAAAAAUGC .........---.............(.((.((((...(((((.((((...(((((...((((((((.(((....))).))))))))))))))))).))))))))).)))........... ( -40.10) >consensus UUCCUAAACCC_CCCCAUAAAAUGAGACUCUGUUACAACGCCUCUG__________________CUUGCAGCGCUGCCGGCGUCGCUGACGUCAGCGGCGUAACAAAGCAAUA_AAUUGC .........................(.((.((((((...(((........(((((....)))))...(((....))).)))(((((((....))))))))))))).)))........... (-23.45 = -24.45 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:14 2006