
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,498,147 – 3,498,247 |
| Length | 100 |
| Max. P | 0.995559 |

| Location | 3,498,147 – 3,498,247 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -21.29 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3498147 100 + 22224390 AAUUUAAAUUAAUUGCCAUAGCCGAGCGUACUGUACUUACUG----UACGCACUUUGCUUGGCA---ACAACAGCACGAAUCGAAAUAUUCCUGCAUAUCCUGCAUA ............((((((.(((.((((((((.((....)).)----)))))..)).))))))))---).....(((.((((......)))).)))............ ( -25.10) >DroSec_CAF1 6765 99 + 1 AAUUUAAAUUAAUUGCCAUAGCCGAGGGGACUGUACUUA-UG----UACGCUCUUCCCUUGGCA---GCAACUGCAGGACCCGAAAUAUUCCUGCAUAUCCUGCAUA ....................((((((((((.(((((...-.)----))))...)))))))))).---(((..(((((((..........))))))).....)))... ( -33.60) >DroSim_CAF1 7173 96 + 1 AAUUUAAAUUAAUUGCCAUAGCCGAGGGGACUGUACUUA-UG----U---CUCUUCCCUUGGCA---GCAACUGCAGGACCCGAAAUAUUCCUGCAUAUCCUGCAUA ....................((((((((((..(.((...-.)----)---)..)))))))))).---(((..(((((((..........))))))).....)))... ( -30.50) >DroEre_CAF1 7188 106 + 1 AAUUUAAAUUAAUUGCCAUAGCCAAGGGCACUGUACUUA-UGUAUGUACGUUAUACCCUGGGCAGGACCAACUGCAGAACCCAAAAUAUUCCUGCAUAUCCUGCAUA .....................(((.(((...(((((...-.....))))).....))))))((((((.....(((((..............)))))..))))))... ( -25.54) >DroYak_CAF1 4015 99 + 1 AAUUUAAAUUAAUUGCCAUAGCCAAGGGAACUGUACUUA-UG----UACGUUCUUCCCCUGGCA---ACAACUGCAGGACCCAAAAUAUUCCUGCAUAUCCUGCAUA .............(((....((((.(((((.(((((...-.)----))))...))))).)))).---.....(((((((..........)))))))......))).. ( -30.20) >consensus AAUUUAAAUUAAUUGCCAUAGCCGAGGGGACUGUACUUA_UG____UACGCUCUUCCCUUGGCA___ACAACUGCAGGACCCGAAAUAUUCCUGCAUAUCCUGCAUA .............(((....((((((((((..(.................)..))))))))))..........((((((..........)))))).......))).. (-21.29 = -21.57 + 0.28)

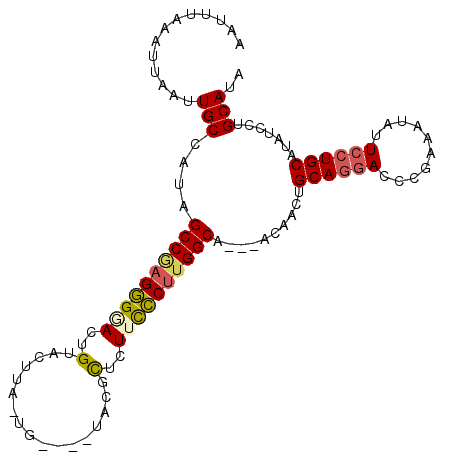

| Location | 3,498,147 – 3,498,247 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -33.52 |
| Consensus MFE | -22.07 |
| Energy contribution | -22.03 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3498147 100 - 22224390 UAUGCAGGAUAUGCAGGAAUAUUUCGAUUCGUGCUGUUGU---UGCCAAGCAAAGUGCGUA----CAGUAAGUACAGUACGCUCGGCUAUGGCAAUUAAUUUAAAUU .......((((.(((.((((......)))).)))))))((---((((((((...(.(((((----(.((....)).)))))).).))).)))))))........... ( -31.60) >DroSec_CAF1 6765 99 - 1 UAUGCAGGAUAUGCAGGAAUAUUUCGGGUCCUGCAGUUGC---UGCCAAGGGAAGAGCGUA----CA-UAAGUACAGUCCCCUCGGCUAUGGCAAUUAAUUUAAAUU ...(((((((.....(((....)))..)))))))((((((---((((.((((..((..(((----(.-...))))..)))))).)))...))))))).......... ( -32.80) >DroSim_CAF1 7173 96 - 1 UAUGCAGGAUAUGCAGGAAUAUUUCGGGUCCUGCAGUUGC---UGCCAAGGGAAGAG---A----CA-UAAGUACAGUCCCCUCGGCUAUGGCAAUUAAUUUAAAUU ...(((((((.....(((....)))..)))))))((((((---((((.((((....(---(----(.-........))))))).)))...))))))).......... ( -29.40) >DroEre_CAF1 7188 106 - 1 UAUGCAGGAUAUGCAGGAAUAUUUUGGGUUCUGCAGUUGGUCCUGCCCAGGGUAUAACGUACAUACA-UAAGUACAGUGCCCUUGGCUAUGGCAAUUAAUUUAAAUU ..((((((((.(((((((..........)))))))....)))))(((.(((((((...((((.....-...)))).))))))).)))....)))............. ( -35.30) >DroYak_CAF1 4015 99 - 1 UAUGCAGGAUAUGCAGGAAUAUUUUGGGUCCUGCAGUUGU---UGCCAGGGGAAGAACGUA----CA-UAAGUACAGUUCCCUUGGCUAUGGCAAUUAAUUUAAAUU ..((((((((...(((((...))))).))))))))((..(---.((((((((((....(((----(.-...))))..)))))))))).)..)).............. ( -38.50) >consensus UAUGCAGGAUAUGCAGGAAUAUUUCGGGUCCUGCAGUUGC___UGCCAAGGGAAGAGCGUA____CA_UAAGUACAGUCCCCUCGGCUAUGGCAAUUAAUUUAAAUU ...(((((((.....(((....)))..)))))))((((((....(((.((((...........................)))).)))....)))))).......... (-22.07 = -22.03 + -0.04)

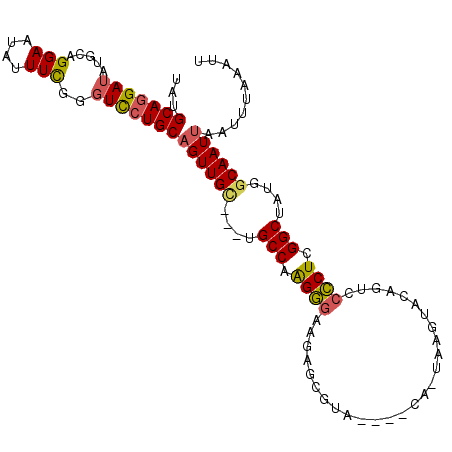
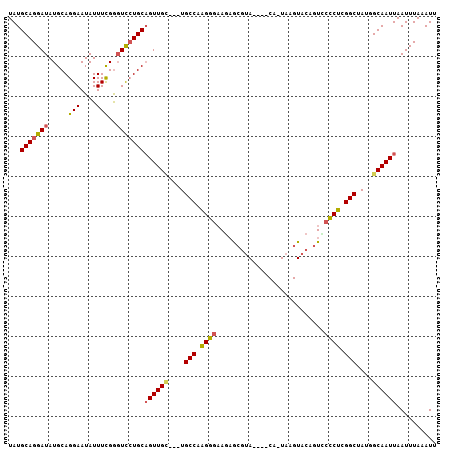
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:03:03 2006