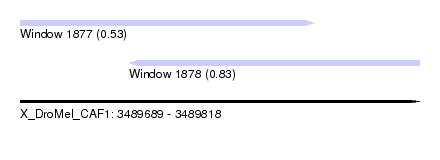
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,489,689 – 3,489,818 |
| Length | 129 |
| Max. P | 0.825904 |

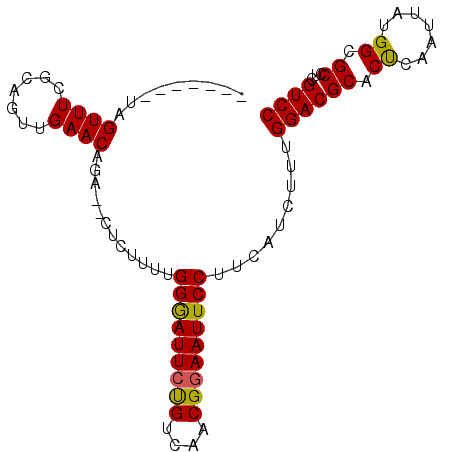
| Location | 3,489,689 – 3,489,784 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -17.26 |
| Energy contribution | -16.93 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3489689 95 + 22224390 CCAACUAUAGUUUCACCAUUGAACAGACACUCUUUUGGAAUUCUGUCAACGAAAUUCCUACAUCUCUGGACGCACUCAAUUAUGGCGCCUUGUCC .........((((.......)))).((((.......((((((.((....)).)))))).........((.(((.(........)))))).)))). ( -15.30) >DroSec_CAF1 50253 86 + 1 -------CAGUUUCGCAGUUGAACAGA--CUCUUUUGGGAUUCUGUCAACGGAAUUCCUUCAUCUUUGGACGCUCCUAAUUAUGGCGCCUCGUCC -------..........(((((.((((--.(((....))).)))))))))((....)).........((((((.((.......)).))...)))) ( -21.10) >DroSim_CAF1 44667 86 + 1 -------UAGUUUCGCAGUUGAACAGA--CUCUUUUGGGAUUCCGUCAACGGAAUUCCUUCAUCUUUGGACGCACUCAAUUAUGGCGCCUCGUCC -------..((((.......)))).((--(......(((((((((....))))))))).........((.(((.(........))))))..))). ( -22.40) >consensus _______UAGUUUCGCAGUUGAACAGA__CUCUUUUGGGAUUCUGUCAACGGAAUUCCUUCAUCUUUGGACGCACUCAAUUAUGGCGCCUCGUCC .........((((.......))))............(((((((((....))))))))).........((((((.((.......)).))...)))) (-17.26 = -16.93 + -0.33)

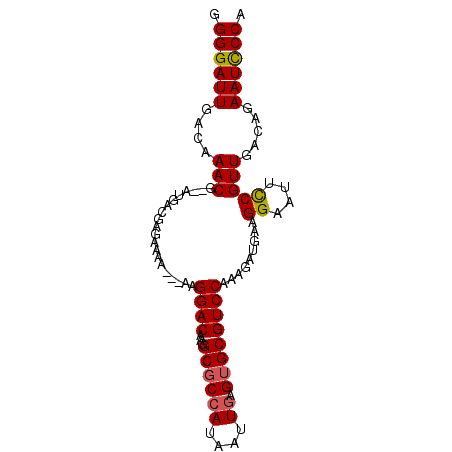
| Location | 3,489,724 – 3,489,818 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -23.69 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3489724 94 - 22224390 GGGGAUUGACAAACGAGAUGACGAGAAAAGCGAAGGACAAGGCGCCAUAAUUGAGUGCGUCCAGAGAUGUAGGAAUUUCGUUGACAGAAUUCCA .((((((....((((((((..(........(...((((...((((((....)).)))))))).).......)..)))))))).....)))))). ( -24.66) >DroSec_CAF1 50279 89 - 1 GGGGAUUGACAAACG--AUGACGAGAAAA---AAGGACGAGGCGCCAUAAUUAGGAGCGUCCAAAGAUGAAGGAAUUCCGUUGACAGAAUCCCA .((((((....(((.--............---..(((((.....((.......))..))))).........((....))))).....)))))). ( -21.30) >DroSim_CAF1 44693 89 - 1 GGGGAUUGACAAACG--AUGACGAGAAAA---AAGGACGAGGCGCCAUAAUUGAGUGCGUCCAAAGAUGAAGGAAUUCCGUUGACGGAAUCCCA .((((..........--............---..((((...((((((....)).)))))))).............(((((....))))))))). ( -23.20) >DroEre_CAF1 53267 89 - 1 GGGGAUUGACAAACG--AUGGGGAGAAAA---AUGGACAAGGCGCCAUAAUUGAGUGCGUCCAAAGAUGCAGGAAUUCCGUUGACUGAAUCCCA .((((((..((..((--((((((......---((((........)))).......((((((....))))))....))))))))..)))))))). ( -25.60) >consensus GGGGAUUGACAAACG__AUGACGAGAAAA___AAGGACAAGGCGCCAUAAUUGAGUGCGUCCAAAGAUGAAGGAAUUCCGUUGACAGAAUCCCA .((((((....(((....................((((...((((((....)).)))))))).........((....))))).....)))))). (-18.58 = -18.70 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:59 2006