| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,483,220 – 3,483,319 |
| Length | 99 |
| Max. P | 0.976577 |

| Location | 3,483,220 – 3,483,319 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -20.71 |
| Energy contribution | -20.52 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
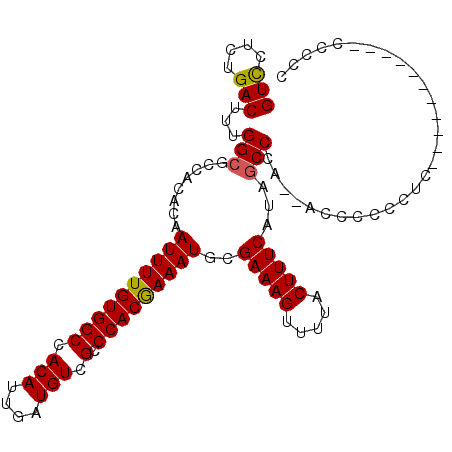
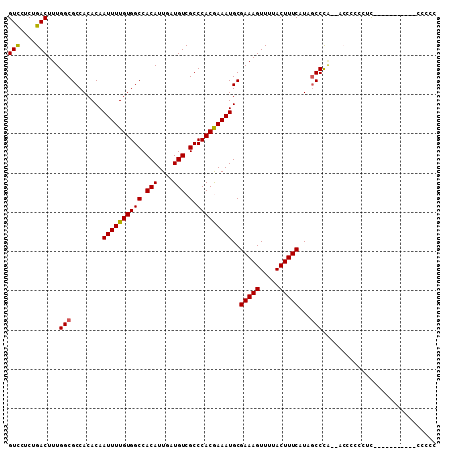
>X_DroMel_CAF1 3483220 99 - 22224390 GUCCUCUGACUUUGGCGCCACACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUAGCCCAUCACCCCCCUCC----------CCCCC (((....)))..(((.((......((((((((((.(((....))).).)))))))))..(((((.....)))))...)))))............----------..... ( -21.80) >DroSec_CAF1 44003 97 - 1 GUCCUCUGACUUUGGCGCCACACAAUUUUGUGGCCACAUUGAUGUCGCCCACAAAAUGCGAAAGUUUUACUUUCAUAGCCCG--ACCCCCCAUA----------CCCCC (((....))).((((.((......((((((((((.(((....))).).)))))))))..(((((.....)))))...)))))--).........----------..... ( -22.60) >DroSim_CAF1 38430 97 - 1 GUCCCCUGACUUUGGCGCCACACAAUUUUGUGGCCACAUUGAUGUCGCCCACAAAAUGCGAAAGUUUUACUUUCAUAGCCCG--ACCCCCCACA----------CCCCC (((....))).((((.((......((((((((((.(((....))).).)))))))))..(((((.....)))))...)))))--).........----------..... ( -22.60) >DroEre_CAF1 46974 88 - 1 GUCCUCUGACUUUGGCGCCACACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUCACCCA-------CUUU--------------CC (((....)))..(((.((((((......)))))).....((((.(((....))).....(((((.....))))))))).)))-------....--------------.. ( -18.80) >DroYak_CAF1 45826 88 - 1 GUCCUCUGACUUUGGCGCCACACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUCGCCCA-------CUUU--------------CC (((....)))...((((.......((((((((((.(((....))).).)))))))))..(((((.....)))))..))))..-------....--------------.. ( -22.60) >DroAna_CAF1 78376 107 - 1 GUUCCCUGACUUUGGCGCCACACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUCGCCUU--GCCCCACUCCCCUGAGCCCUUCCCC .............((((.......((((((((((.(((....))).).)))))))))..(((((.....)))))..))))..--......(((....)))......... ( -22.60) >consensus GUCCUCUGACUUUGGCGCCACACAAUUUUGUGGCCACAUUGAUGUCGCCCACGAAAUGCGAAAGUUUUACUUUCAUAGCCCA__ACCCCCCUC___________CCCCC (((....)))...(((........((((((((((.(((....))).).)))))))))..(((((.....)))))...)))............................. (-20.71 = -20.52 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:55 2006