| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,476,503 – 3,476,600 |
| Length | 97 |
| Max. P | 0.877854 |

| Location | 3,476,503 – 3,476,600 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 72.61 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.03 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
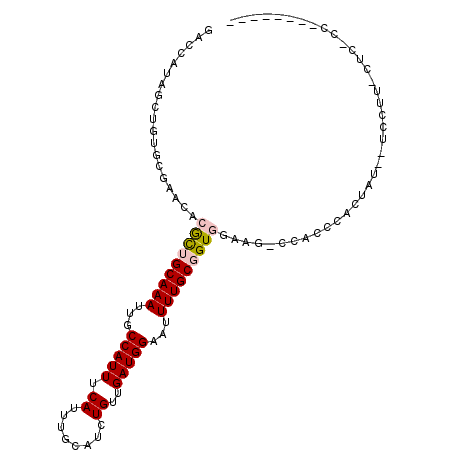
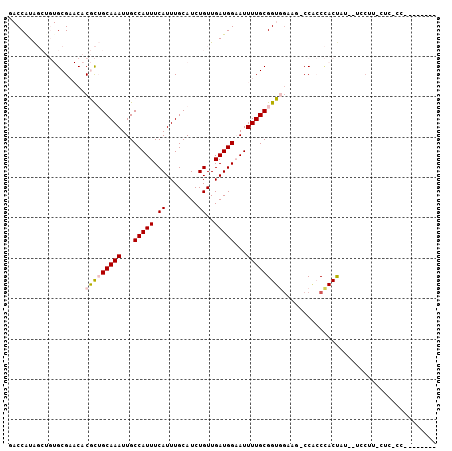
>X_DroMel_CAF1 3476503 97 + 22224390 GACCGUAGCUGUGCGAACACGCUGCAAAUUGCCAUUUCAUUUGCAUCUGUUGAUGGAAUUUUGCGGUGGAAG-CCGCCCACCCUCCUCCUUCCUCCCC-------- ....((((((((....))).))))).....((.((((((((.((....)).))))))))...))(((((...-....)))))................-------- ( -25.00) >DroPse_CAF1 45582 96 + 1 --CCAUG-----GCG--CACAUGGCAAAUUGCCAUUUCAUUUGCAUCUGUUGAUGGCAUUUUGCGGUUGCAU-CCAACAACUAUGUUGCCUUCUCAUCCUCUGAGU --....(-----(((--.((((((((((.(((((((.((........))..))))))).))))).((((...-.))))...)))))))))..((((.....)))). ( -28.90) >DroSec_CAF1 37596 89 + 1 GACCGUAGCUGUGCGAACACGCUGCAAAUUGCCAUUUCAUUUGCAUCUGUUGAUGGAAUUUUGCGGUGGAAG-CCGCCCACUACCCUCCU---------------- ((..((((.((.(((....(((((((((...(((((.((........))..)))))...)))))))))....-.))).))))))..))..---------------- ( -27.30) >DroEre_CAF1 40326 95 + 1 GACCAUAGCUGUGCGAACACGCUGCAAAUUGCCAUUUCAUUUGCAUCUGUUGAUGGAAUUUUGCGGUGGAAG-CCACCCACUCC--CACUUCCACUCC-------- ......((((((....))).)))(((((...(((((.((........))..)))))...)))))((((((((-...........--..))))))))..-------- ( -24.32) >DroYak_CAF1 39298 84 + 1 GACCAUAGCUGUGCGAACACGCUGCAAAUUGCCAUUUCAUUUGCAUCUGUUGAUGGAAUUUUGCGGUGAAAAACCUCCCACU--------------CC-------- ..........(((.((...(((((((((...(((((.((........))..)))))...))))))))).......)).))).--------------..-------- ( -18.60) >DroPer_CAF1 47109 96 + 1 --CCAUG-----GCG--CACAUGGCAAAUUGCCAUUUCAUUUGCAUCUGUUGAUGGCAUUUUGCGGUUGCAU-CCAACUACUUUGUUGCCUUCUCACCCUCGGAGU --....(-----(((--((.((.(((((.(((((((.((........))..))))))).))))).)))))..-..(((......))))))..(((.......))). ( -27.10) >consensus GACCAUAGCUGUGCGAACACGCUGCAAAUUGCCAUUUCAUUUGCAUCUGUUGAUGGAAUUUUGCGGUGGAAG_CCACCCACUAU__UCCUU_CUC_CC________ ...................(((((((((...(((((.((........))..)))))...)))))))))...................................... (-14.81 = -15.03 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:52 2006