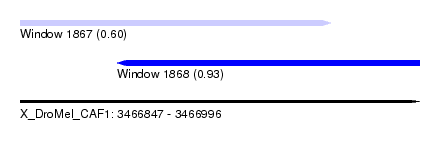
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,466,847 – 3,466,996 |
| Length | 149 |
| Max. P | 0.934713 |

| Location | 3,466,847 – 3,466,963 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -15.14 |
| Energy contribution | -15.50 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3466847 116 + 22224390 CGCAUUUCCCCCAAACGAACUUAUCCCGAGUCCCAAAUUUGUGAAAAGUAAUAACAAAAGGCGGCCAAAAGAAGCCGCCAAAAUACGUUUGAAUUGCUCACAAAUAAUAAAGAAAU .(((.(((....(((((.((((.((.(((((.....))))).)).))))..........((((((........))))))......)))))))).)))................... ( -26.70) >DroSec_CAF1 27095 113 + 1 CGCAUUUCCCCCA-CCAAACUUAUC-CGAGUCCCAAAA-UGUGAAAAGUAGUAACAAAAGGCGGGCAAAAGAAGCCGCCAAAAUACGAUUGAAUUUCCCACAAAUAAUAAAGAAAU ...(((((.....-....((((...-.)))).......-((((....((....))....(((((.(.......))))))...................)))).........))))) ( -14.00) >DroSim_CAF1 26875 113 + 1 CGCAUUUCCCCCA-CCACACUUAUC-CGAGUCCCAAAA-UGUGAAAAGUAGUAACAAAAGGCGGCCAAAAGAAGCCGCCAAAAUACGAUUGAAUUGCCCACAAAUAAUAAAGAAAU .(((.(((.....-..((((((.((-...((......)-)..)).)))).)).......((((((........))))))...........))).)))................... ( -19.20) >DroEre_CAF1 30463 113 + 1 CGCAUUUCCUCCA-CCGAACUUAUC-CAACUCCCAAAU-UGUGAAAAGUAGUAACAAAAGACGGCCAAAAGAAGCCGCCAAACUACGCUUGAAUUGCACACAAAUAAUAAAGAAAU .(((.(((.....-....((((.((-(((........)-)).)).))))..........(.((((........)))).)...........))).)))................... ( -15.20) >DroYak_CAF1 28575 111 + 1 CGCAUUUCC-CCA-CCGAACUUAUC-CAAGUCCCAAAA-UGUGAAAAGUAGUAACAAAAGGCGGCCAA-AGAAGCCGCCAAACUACGCUUGAAUUGCCCACAAAUAAUAAAGAAAU ...(((((.-...-..(.((((...-.)))).).....-((((....(((((.......((((((...-....))))))..)))))((.......)).)))).........))))) ( -20.80) >consensus CGCAUUUCCCCCA_CCGAACUUAUC_CGAGUCCCAAAA_UGUGAAAAGUAGUAACAAAAGGCGGCCAAAAGAAGCCGCCAAAAUACGAUUGAAUUGCCCACAAAUAAUAAAGAAAU .(((.(((..........((((.((.((...........)).)).))))..........((((((........))))))...........))).)))................... (-15.14 = -15.50 + 0.36)

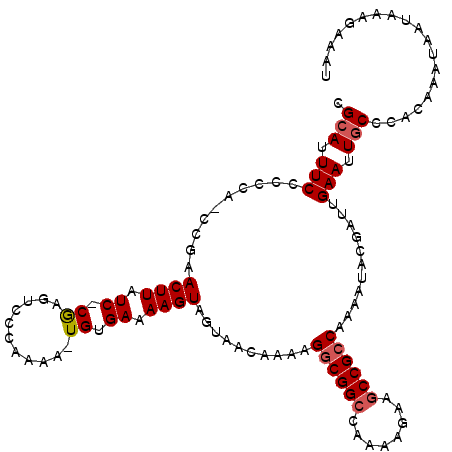
| Location | 3,466,883 – 3,466,996 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.40 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -10.64 |
| Energy contribution | -11.67 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3466883 113 - 22224390 -------ACAAAGUGAAAUAGGCAAUGCUCAAAUUGAAUUAUUUCUUUAUUAUUUGUGAGCAAUUCAAACGUAUUUUGGCGGCUUCUUUUGGCCGCCUUUUGUUAUUACUUUUCACAAAU -------.....((((((.((....((((((.....(((.........))).....)))))).....((((......(((((((......)))))))...))))....)))))))).... ( -29.30) >DroSec_CAF1 27129 101 - 1 -------ACAAAGGG-----------GCUCAAAUUGAAUUAUUUCUUUAUUAUUUGUGGGAAAUUCAAUCGUAUUUUGGCGGCUUCUUUUGCCCGCCUUUUGUUACUACUUUUCACA-UU -------((((((((-----------(((.(((((((...(((((((..........)))))))....))).)))).)))(((.......)))..))))))))..............-.. ( -21.00) >DroSim_CAF1 26909 101 - 1 -------ACAAAGUG-----------GCUCAAAUUGAAUUAUUUCUUUAUUAUUUGUGGGCAAUUCAAUCGUAUUUUGGCGGCUUCUUUUGGCCGCCUUUUGUUACUACUUUUCACA-UU -------..((((((-----------(..(((((((((((...(((.((.....)).))).))))))).........(((((((......))))))).))))...))))))).....-.. ( -24.60) >DroEre_CAF1 30497 117 - 1 AAACAAAAAAAGGG--AAUAGGCAAUGUUGAAAUUGAAUUAUUUCUUUAUUAUUUGUGUGCAAUUCAAGCGUAGUUUGGCGGCUUCUUUUGGCCGUCUUUUGUUACUACUUUUCACA-AU .........(((((--(((((.((((......))))..)))))))))).....(((((.((.......))(((((..(((((((......))))))).......)))))....))))-). ( -28.50) >DroYak_CAF1 28608 109 - 1 -------AAAAAGG--AAUAGGCAAUGUUCAAAUUGAAUUAUUUCUUUAUUAUUUGUGGGCAAUUCAAGCGUAGUUUGGCGGCUUCU-UUGGCCGCCUUUUGUUACUACUUUUCACA-UU -------..(((((--(((((.((((......))))..))))))))))......(((((((.......))(((((..(((((((...-..))))))).......)))))...)))))-.. ( -31.70) >DroAna_CAF1 50995 94 - 1 -------AUUAAGCA-AAAAGGCGGAAUGGAAAUUAAAUUACAUCCUUAAUGAUUUUUUUUGUUUCA--CGUAGUUUGGCGGUUUCUU---------------UACCACUUUUCGGA-UU -------......(.-.(((((.((...((((((((((((((......((..(......)..))...--.))))))))...)))))).---------------..)).)))))..).-.. ( -14.30) >consensus _______ACAAAGGG_AAUAGGCAAUGCUCAAAUUGAAUUAUUUCUUUAUUAUUUGUGGGCAAUUCAAGCGUAGUUUGGCGGCUUCUUUUGGCCGCCUUUUGUUACUACUUUUCACA_UU .........((((.............(((((.....(((.........))).....))))).........(((....(((((((......)))))))......)))..))))........ (-10.64 = -11.67 + 1.03)
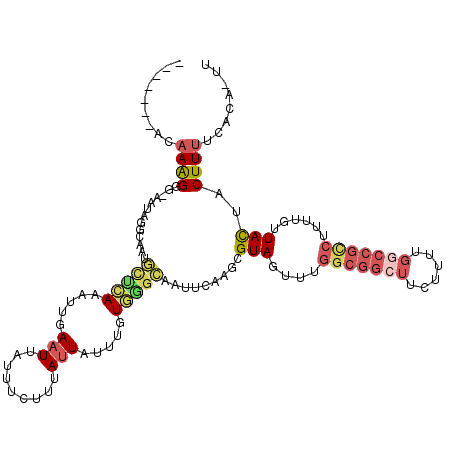
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:50 2006