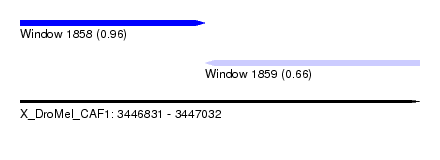
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,446,831 – 3,447,032 |
| Length | 201 |
| Max. P | 0.959296 |

| Location | 3,446,831 – 3,446,924 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -11.27 |
| Energy contribution | -11.51 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3446831 93 + 22224390 AUUCCGCCCGCUUUUCGUUUCCAAUCCGAAUCAUUUUCGGGCACUUAAAAAGGGAAGAAUUUCUAGCUUUUCCCACCCCUU-------UUUUUUCUUCAA .....(((((...((((.........)))).......))))).........(((((((.........))))))).......-------............ ( -18.00) >DroSec_CAF1 19019 89 + 1 AUU-UGCCCGCUUUUCGUUUCCAAUCCGAAUCAUUUU-GGGCACUUAU-AAGGGAAGGUUUUCCAGCUUUUCCCACCCCUU-------C-UCUUCUUCAA ...-((((((...((((.........))))......)-))))).....-(((((((((...................))))-------)-))))...... ( -18.31) >DroSim_CAF1 19119 90 + 1 AUU-UGCCCGCUUUUCGUUUCCAAUCCGAAUCAUUUU-GGGCACUUUU-AAGGGAAGGUUUUCCAGCUUUUCCCACCCCUC-------CUUCUUCUUCAA ...-((((((...((((.........))))......)-))))).....-..((((((((......))))))))........-------............ ( -17.80) >DroEre_CAF1 22255 97 + 1 AUU-CGGCCUCUUUUGGUUUUCCAUCCGAAUCAUUUG-GGUCACUUAU-AAGGGAAGGUUCUGUAACUUUUCCCACCCGUUAAUAAUUCCUCUUUUUCAA ...-.(((((....((((((.......))))))...)-))))......-..(((((((((....)))))))))........................... ( -17.60) >DroYak_CAF1 20287 97 + 1 AUU-UGCCCGCUUUUGGUUACCCAUCCGAAUCAUUUC-GGUCUCUUAU-AGGGGAAGGUUUUUUAACUUUUCCCACCCGUUCAUAACUCCUCUUCUUCAA ...-...........(((((.....(((((....)))-))........-.((((((((((....)))))))))).........)))))............ ( -20.40) >consensus AUU_UGCCCGCUUUUCGUUUCCAAUCCGAAUCAUUUU_GGGCACUUAU_AAGGGAAGGUUUUCUAGCUUUUCCCACCCCUU_______CCUCUUCUUCAA .....((((.........(((......)))........)))).........(((((((((....)))))))))........................... (-11.27 = -11.51 + 0.24)

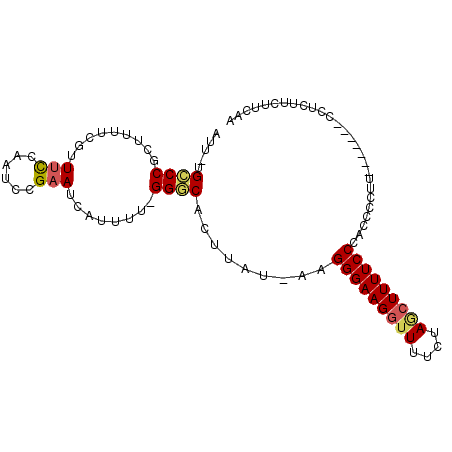
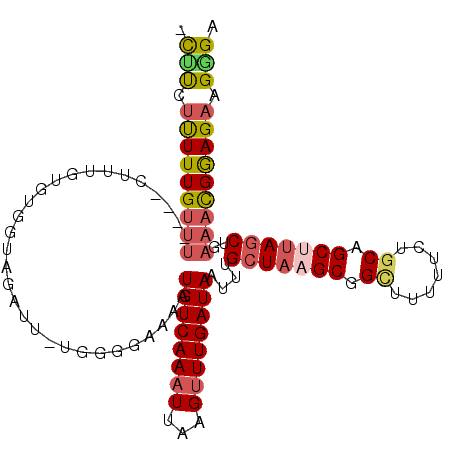
| Location | 3,446,924 – 3,447,032 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 84.03 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -19.03 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3446924 108 - 22224390 -CUUCUUUUUGUUUCUUUCUUUGUGUGGUAGAUU-UGGGGAAAAUGUCAAAUUAAGUUUGAUAAUUUGCUAAGCGGCUUUUCCUGCAGCUUAGCUGAAACGGAGAAGGGA -((..(((((((((((((((..(.((.....)).-)..))))).((((((((...))))))))....(((((((.((.......)).))))))).))))))))))..)). ( -32.20) >DroSec_CAF1 19108 104 - 1 -CCUCUUUUUGUUU----CUUUGUGUGGUAGAUU-UGGCGAAAAUGUCAAAUUAAGUUUGAUAAUUAGCUAAGCGGUUUUUUCUGCAGCAUAGCUGAAACGGAGAAGGGA -((..(((((((((----(.((((((.(((((((-((((.....((((((((...))))))))....))))))........))))).))))))..))))))))))..)). ( -32.80) >DroSim_CAF1 19209 104 - 1 -CCUCUUUUUGUUU----CUUUGUGUGGUAGAUU-UGGCGAAAAUGUCAAAUUAAGUUUGAUAAUUUGCUAAGCGGCUUUUUCUGCAGCUUAGCUGAAACGGAGAAGGGA -((..(((((((((----(...........((((-((((......))))))))..............(((((((.((.......)).))))))).))))))))))..)). ( -33.30) >DroEre_CAF1 22352 105 - 1 CUGCUUUUUUGUUU----CUUUGUGCGGCAGAUU-UGGGGAAAAUGUCAAAUUAAGUUUGAUAAUUUGCUAAGCGGCUUUUUCGGCGGCUUAGCUGAAAUGGAGAAGGGA ...((((((..(((----(.........((((((-(((.((.....))...))))))))).......(((((((.(((.....))).))))))).))))..))))))... ( -32.40) >DroYak_CAF1 20384 104 - 1 -UUUCUUUUUGUUU----CUUUGUGUGGUAGAUU-UGCGAAAAAUGUCAAAUUAAGAUUGAUAAUUUGCUAAGCGGCUUUUUCUGCGGCUUAGCCGAAAUGGAGAAGGGA -(..(((((..(((----(....((..(.....)-..)).....((((((.......))))))....(((((((.((.......)).))))))).))))..))).))..) ( -26.70) >DroAna_CAF1 33333 99 - 1 -UUUCGCUUUGAU-----UUUUGUGUGGGAGCUUCUUAGCCAAAUGUCAAAUUAUGUUUGAUAAUUUGCUAAGCGGCUUU-----CGGCAAAUCUGUCAUGAAGAAGAGA -((((....((((-----.(((((.((..((((.((((((.((.((((((((...)))))))).)).)))))).))))..-----)))))))...))))....))))... ( -32.50) >consensus _CUUCUUUUUGUUU____CUUUGUGUGGUAGAUU_UGGGGAAAAUGUCAAAUUAAGUUUGAUAAUUUGCUAAGCGGCUUUUUCUGCAGCUUAGCUGAAACGGAGAAGGGA .(((.(((((((((..............................((((((((...))))))))....(((((((.((.......)).)))))))..))))))))).))). (-19.03 = -19.57 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:42 2006