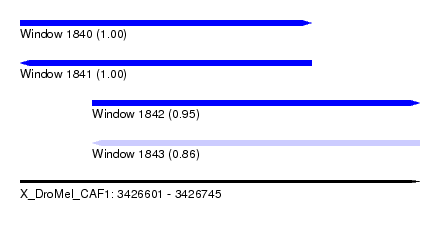
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,426,601 – 3,426,745 |
| Length | 144 |
| Max. P | 0.998328 |

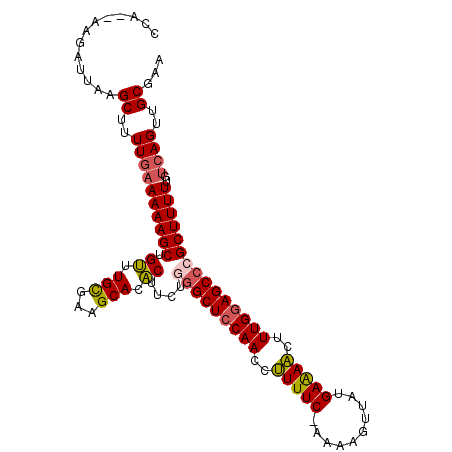
| Location | 3,426,601 – 3,426,706 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3426601 105 + 22224390 CCA--UAGAUGAAGCUUUUGAAAAAGCUGUUUGCGUAGCAUACUUCUGGGCUCCAACCUUUUC-AAAAGUUAUGAAAACUUUGUAGCCCGCUUUUUGUCAGUUGCGAA ...--........((..((((((((((.((.(((...))).))....(((((.(((..(((((-(.......))))))..))).)))))))))))..))))..))... ( -28.40) >DroSec_CAF1 2021 105 + 1 CCA--CAGAUUAAGCUUUUGAAAAAGCUGUUUGCGAAGCACACUUCUAGGCUCCAACCCUUUC-AAAAGUUAUGAAAGCUUUGGAGCCCGCUUUUUGUCAGUUGCGAA ...--........((..((((((((((.......((((....))))..((((((((..(((((-(.......))))))..)))))))).))))))..))))..))... ( -33.70) >DroSim_CAF1 2019 105 + 1 CCA--CAGAUUAAGCUUUUGAAAAAGCUGUUUGCGUAGCACACUUCUAGGCUCCAACCCUUUC-AAAAGUUAUGAAAGCUUUGGAGCCCGCUUUUAGGCAGUUGCGAA ...--(((((..((((((....)))))))))))((((((.........((((((((..(((((-(.......))))))..)))))))).(((....))).)))))).. ( -36.40) >DroEre_CAF1 594 107 + 1 CCAGUAAGAUUAAGCUUUUGAAAAAGCUGACUGUGAAGCACUCUUUUGGGCUCCAACCUUUUC-UAAGUGUAUGAGAACUUUGGAGCACGCUUUUUGUCAGUUGCGAA ................((((....(((((((...(((((.(......).(((((((..(((((-.........)))))..)))))))..)))))..))))))).)))) ( -29.80) >DroYak_CAF1 690 108 + 1 CCAGUAAGAUUAAGCUUUUUAAAAAGCUGUUUGCGAAGCACACUUUCGGGCUCCAACCUUUUCCCAAAAGCAUGAGAACUUUGGAGCCCGCUUUUUAUCAGUUGCGAA ............((((...((((((((.((.(((...))).))....(((((((((..(((((..........)))))..)))))))))))))))))..))))..... ( -31.60) >consensus CCA__AAGAUUAAGCUUUUGAAAAAGCUGUUUGCGAAGCACACUUCUGGGCUCCAACCUUUUC_AAAAGUUAUGAAAACUUUGGAGCCCGCUUUUUGUCAGUUGCGAA .............((..((((((((((.((.(((...))).))....(((((((((..(((((..........)))))..)))))))))))))))..))))..))... (-26.74 = -26.74 + 0.00)

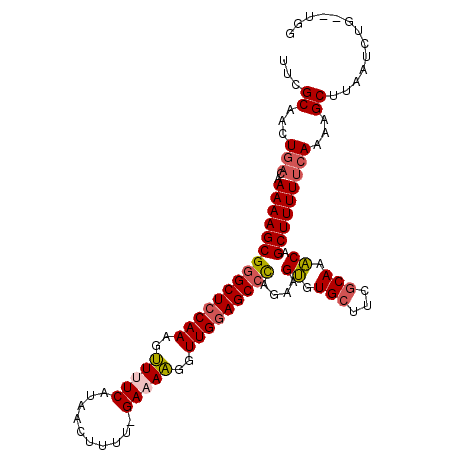
| Location | 3,426,601 – 3,426,706 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -26.48 |
| Energy contribution | -26.84 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3426601 105 - 22224390 UUCGCAACUGACAAAAAGCGGGCUACAAAGUUUUCAUAACUUUU-GAAAAGGUUGGAGCCCAGAAGUAUGCUACGCAAACAGCUUUUUCAAAAGCUUCAUCUA--UGG ...((...(((..(((((((((((.(((..((((((.......)-)))))..))).)))))....((.(((...))).)).)))))))))...))........--... ( -28.20) >DroSec_CAF1 2021 105 - 1 UUCGCAACUGACAAAAAGCGGGCUCCAAAGCUUUCAUAACUUUU-GAAAGGGUUGGAGCCUAGAAGUGUGCUUCGCAAACAGCUUUUUCAAAAGCUUAAUCUG--UGG ..((((...........(((((((((((..((((((.......)-)))))..))))))))).((((....))))))....((((((....)))))).....))--)). ( -37.00) >DroSim_CAF1 2019 105 - 1 UUCGCAACUGCCUAAAAGCGGGCUCCAAAGCUUUCAUAACUUUU-GAAAGGGUUGGAGCCUAGAAGUGUGCUACGCAAACAGCUUUUUCAAAAGCUUAAUCUG--UGG ...((((((((......))(((((((((..((((((.......)-)))))..)))))))))...))).)))..((((...((((((....)))))).....))--)). ( -35.40) >DroEre_CAF1 594 107 - 1 UUCGCAACUGACAAAAAGCGUGCUCCAAAGUUCUCAUACACUUA-GAAAAGGUUGGAGCCCAAAAGAGUGCUUCACAGUCAGCUUUUUCAAAAGCUUAAUCUUACUGG ...((...(((..(((((((.(((((((..((((.........)-)))....))))))).)....((.((.....)).)).)))))))))...))............. ( -23.20) >DroYak_CAF1 690 108 - 1 UUCGCAACUGAUAAAAAGCGGGCUCCAAAGUUCUCAUGCUUUUGGGAAAAGGUUGGAGCCCGAAAGUGUGCUUCGCAAACAGCUUUUUAAAAAGCUUAAUCUUACUGG ...((......(((((((((((((((((..((((((......))))))....)))))))))....((.(((...))).)).))))))))....))............. ( -35.90) >consensus UUCGCAACUGACAAAAAGCGGGCUCCAAAGUUUUCAUAACUUUU_GAAAAGGUUGGAGCCCAGAAGUGUGCUUCGCAAACAGCUUUUUCAAAAGCUUAAUCUG__UGG ...((...(((..(((((((((((((((..(((((..........)))))..)))))))))....((.(((...))).)).)))))))))...))............. (-26.48 = -26.84 + 0.36)

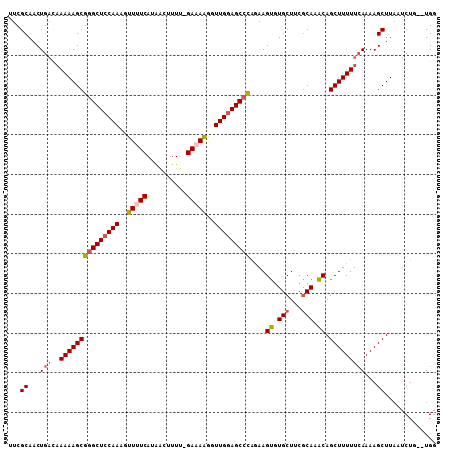
| Location | 3,426,627 – 3,426,745 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -22.79 |
| Energy contribution | -22.23 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3426627 118 + 22224390 GUUUGCGUAGCAUACUUCUGGGCUCCAACCUUUUC-AAAAGUUAUGAAAACUUUGUAGCCCGCUUUUUGUCAGUUGCGAACCCUAGGCUAGGCUAUGACUUUGGUUUCAGCUUUCCA-AA (((((((..(((......((((((.(((..(((((-(.......))))))..))).)))))).....)))....)))))))....((..(((((..(((....)))..))))).)).-.. ( -32.40) >DroSec_CAF1 2047 113 + 1 GUUUGCGAAGCACACUUCUAGGCUCCAACCCUUUC-AAAAGUUAUGAAAGCUUUGGAGCCCGCUUUUUGUCAGUUGCGAACCCUGG-----GCUAUGGCUUUGGUUCCAGCUUUUCA-AA ....((((((....))))..((((((((..(((((-(.......))))))..)))))))).))..((((..(((((.(((((..((-----((....)))).))))))))))...))-)) ( -41.10) >DroSim_CAF1 2045 113 + 1 GUUUGCGUAGCACACUUCUAGGCUCCAACCCUUUC-AAAAGUUAUGAAAGCUUUGGAGCCCGCUUUUAGGCAGUUGCGAACCCUGG-----GCUAUGGCUUUGGUUCCAGCUUUUCA-AA ....(((((((.........((((((((..(((((-(.......))))))..)))))))).(((....))).)))))(((((..((-----((....)))).)))))..))......-.. ( -40.00) >DroEre_CAF1 622 113 + 1 GACUGUGAAGCACUCUUUUGGGCUCCAACCUUUUC-UAAGUGUAUGAGAACUUUGGAGCACGCUUUUUGUCAGUUGCGAAACCAAG-----GCUAUGUCCUUGCUAUCAGCUUUUCA-AA (((...(((((.(......).(((((((..(((((-.........)))))..)))))))..)))))..)))(((((......((((-----(......)))))....))))).....-.. ( -29.90) >DroYak_CAF1 718 115 + 1 GUUUGCGAAGCACACUUUCGGGCUCCAACCUUUUCCCAAAAGCAUGAGAACUUUGGAGCCCGCUUUUUAUCAGUUGCGAACCCUGU-----GCUAUGUCUUUGGUUUUAGCUUUUUAAAA ((((((((..........((((((((((..(((((..........)))))..))))))))))...........)))))))).....-----((((...(....)...))))......... ( -32.70) >consensus GUUUGCGAAGCACACUUCUGGGCUCCAACCUUUUC_AAAAGUUAUGAAAACUUUGGAGCCCGCUUUUUGUCAGUUGCGAACCCUGG_____GCUAUGGCUUUGGUUUCAGCUUUUCA_AA ((.(((...))).))...((((((((((..(((((..........)))))..)))))))))).........(((((.(((((....................))))))))))........ (-22.79 = -22.23 + -0.56)

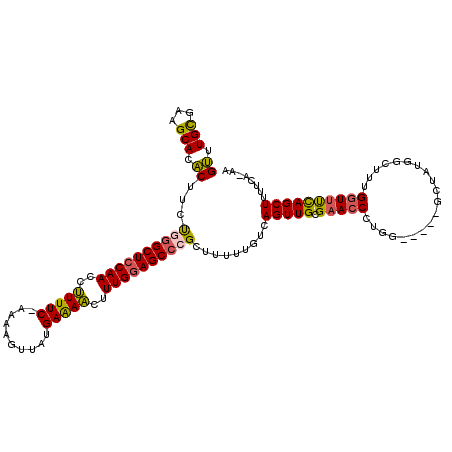
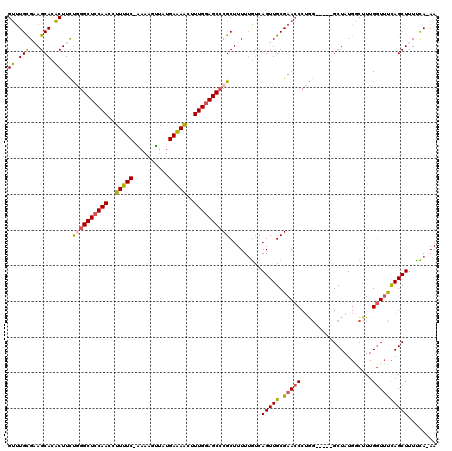
| Location | 3,426,627 – 3,426,745 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -21.37 |
| Energy contribution | -21.45 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3426627 118 - 22224390 UU-UGGAAAGCUGAAACCAAAGUCAUAGCCUAGCCUAGGGUUCGCAACUGACAAAAAGCGGGCUACAAAGUUUUCAUAACUUUU-GAAAAGGUUGGAGCCCAGAAGUAUGCUACGCAAAC ((-(((..........)))))......((.((((....(.(((((............))(((((.(((..((((((.......)-)))))..))).))))).))).)..)))).)).... ( -29.30) >DroSec_CAF1 2047 113 - 1 UU-UGAAAAGCUGGAACCAAAGCCAUAGC-----CCAGGGUUCGCAACUGACAAAAAGCGGGCUCCAAAGCUUUCAUAACUUUU-GAAAGGGUUGGAGCCUAGAAGUGUGCUUCGCAAAC ((-((...((.(((((((...((....))-----....))))).)).))..))))..(((((((((((..((((((.......)-)))))..))))))))).((((....)))))).... ( -40.70) >DroSim_CAF1 2045 113 - 1 UU-UGAAAAGCUGGAACCAAAGCCAUAGC-----CCAGGGUUCGCAACUGCCUAAAAGCGGGCUCCAAAGCUUUCAUAACUUUU-GAAAGGGUUGGAGCCUAGAAGUGUGCUACGCAAAC ..-......((..(((((...((....))-----....)))))((((((((......))(((((((((..((((((.......)-)))))..)))))))))...))).)))...)).... ( -38.10) >DroEre_CAF1 622 113 - 1 UU-UGAAAAGCUGAUAGCAAGGACAUAGC-----CUUGGUUUCGCAACUGACAAAAAGCGUGCUCCAAAGUUCUCAUACACUUA-GAAAAGGUUGGAGCCCAAAAGAGUGCUUCACAGUC ..-((((..(((((.(.(((((......)-----)))).).))................(.(((((((..((((.........)-)))....))))))).).....)))..))))..... ( -25.00) >DroYak_CAF1 718 115 - 1 UUUUAAAAAGCUAAAACCAAAGACAUAGC-----ACAGGGUUCGCAACUGAUAAAAAGCGGGCUCCAAAGUUCUCAUGCUUUUGGGAAAAGGUUGGAGCCCGAAAGUGUGCUUCGCAAAC .....................(....(((-----(((...(((((............))(((((((((..((((((......))))))....))))))))))))..))))))...).... ( -35.80) >consensus UU_UGAAAAGCUGAAACCAAAGACAUAGC_____CCAGGGUUCGCAACUGACAAAAAGCGGGCUCCAAAGUUUUCAUAACUUUU_GAAAAGGUUGGAGCCCAGAAGUGUGCUUCGCAAAC ........(((................((........((........))........))(((((((((..(((((..........)))))..)))))))))........)))........ (-21.37 = -21.45 + 0.08)

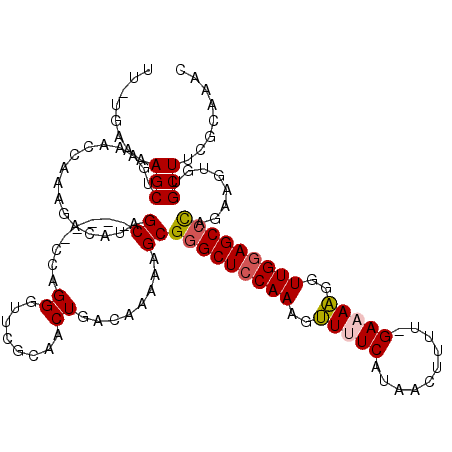
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:28 2006