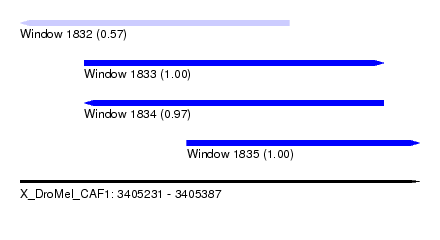
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,405,231 – 3,405,387 |
| Length | 156 |
| Max. P | 0.997995 |

| Location | 3,405,231 – 3,405,336 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -16.85 |
| Energy contribution | -16.80 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3405231 105 - 22224390 GUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGAAGCACUCAUUAGGAAAUCCUCAACUGAUGUUCGCUCCCGCUUACGC---CGC------------ ((((...((((((((((((((.((((....)))).))))..)))))))))).....(((...((((((...........))))))...))).))))......---...------------ ( -26.20) >DroVir_CAF1 74695 102 - 1 GUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGGAGCACUCAUUAGGAAAUCCUCUGCUGCUGCUAGCUAC-ACUUC--G----CCU----------- ((((...((((((((((((((.((((....)))).))))..))))))))))..(((((((..((..(((....))).)).)))).))).))))-.....--.----...----------- ( -24.40) >DroGri_CAF1 71914 101 - 1 GUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGGAGCACUCAUUAGGAAAUCCUCUGCUGCUGC--GCUAC-ACUUG--U----CUUC---------- ((((...((((((((((((((.((((....)))).))))..))))))))))...(.((((..((..(((....))).)).)))).)--.))))-.....--.----....---------- ( -24.80) >DroSec_CAF1 51628 117 - 1 GUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGAAGCACUCAUUAGGAAAUCCUCAACUGAUGUUCGCUCCCGCUUUCGA---CGCCCCCCGCCCACC ((((...((((((((((((((.((((....)))).))))..)))))))))).....(((...((((((...........))))))...))).))))......---............... ( -26.20) >DroEre_CAF1 63310 105 - 1 GUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGAAGCACUCAUUAGGAAAUCCUCAACUGAUGUUCGC-----CUUUCACCCACCCCC---------- ((((...((((((((((((((.((((....)))).))))..))))))))))..((((((...((((((...........))))))...))-----..)))).))))....---------- ( -26.60) >DroWil_CAF1 109354 94 - 1 GUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAUUUCAUUCAUGAAGCACUCAUUAGGAAAUCCUCAACUGCUGCCC-----CACUGU--------------------- ((((...((((((.(((((((.((((....)))).))))..))).)))))).....((((......(((....)))....))))...)-----)))...--------------------- ( -21.00) >consensus GUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGAAGCACUCAUUAGGAAAUCCUCAACUGAUGCUCGCU_C_ACUUU__C____CC____________ (.(((..((((((((((((((.((((....)))).))))..)))))))))).((((.....)))).......))).)........................................... (-16.85 = -16.80 + -0.05)

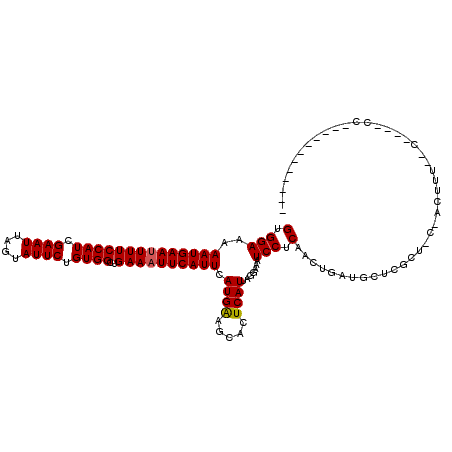
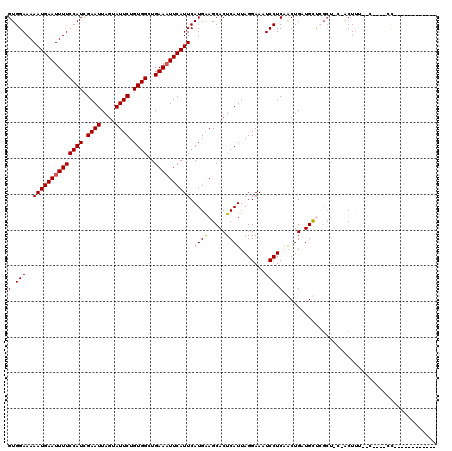
| Location | 3,405,256 – 3,405,373 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 97.44 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
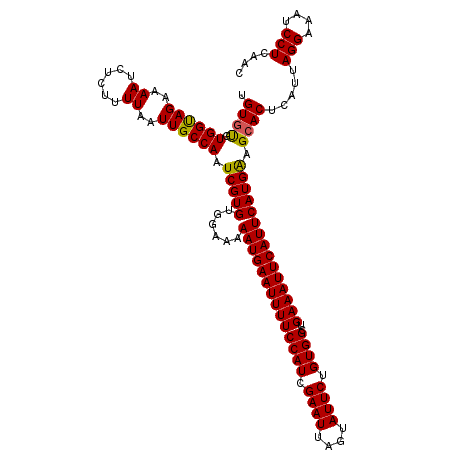
>X_DroMel_CAF1 3405256 117 + 22224390 GUUGAGGAUUUCCUAAUGAGUGCUUCAUGAAUGAAUUUCAGCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACGCACA ((.((((((((((((((..(((......((((((((((...(((..((((....))))..))).)))))))))).....))).))))).........))))))))).))........ ( -25.20) >DroVir_CAF1 74717 117 + 1 GCAGAGGAUUUCCUAAUGAGUGCUCCAUGAAUGAAUUUCAGCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACACACA ..(((((((((((((((..(((......((((((((((...(((..((((....))))..))).)))))))))).....))).))))).........)))))))))).......... ( -25.90) >DroGri_CAF1 71935 117 + 1 GCAGAGGAUUUCCUAAUGAGUGCUCCAUGAAUGAAUUUCAGCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACACACA ..(((((((((((((((..(((......((((((((((...(((..((((....))))..))).)))))))))).....))).))))).........)))))))))).......... ( -25.90) >DroYak_CAF1 54332 117 + 1 GUUGAGGAUUUCCUAAUGAGUGCUUCAUGAAUGAAUUUCAGCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACGCACA ((.((((((((((((((..(((......((((((((((...(((..((((....))))..))).)))))))))).....))).))))).........))))))))).))........ ( -25.20) >DroMoj_CAF1 87704 117 + 1 GCAGAGGAUUUCCUAAUGAGUGCUCCAUGAAUGAAUUUCAGCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUGCCACACACA (((((((((((((((((..(((......((((((((((...(((..((((....))))..))).)))))))))).....))).))))).........))))))))))))........ ( -31.70) >DroAna_CAF1 135514 117 + 1 GUUGAGGAUUUCCUAAUGAGUGCUUCAUGAAUGAAUUUCAGCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACAUACA ((.((((((((((((((..(((......((((((((((...(((..((((....))))..))).)))))))))).....))).))))).........))))))))).))........ ( -25.20) >consensus GCAGAGGAUUUCCUAAUGAGUGCUCCAUGAAUGAAUUUCAGCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACACACA ((.((((((((((((((..(((......((((((((((...(((..((((....))))..))).)))))))))).....))).))))).........))))))))).))........ (-24.62 = -24.78 + 0.17)

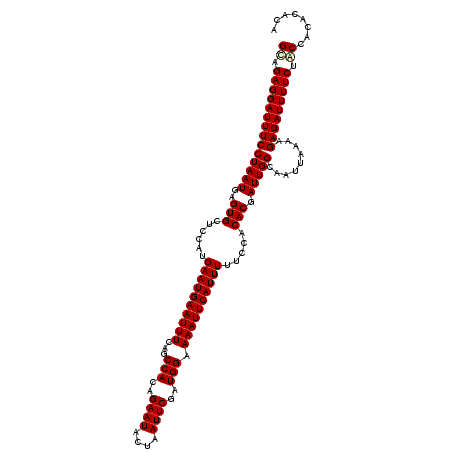
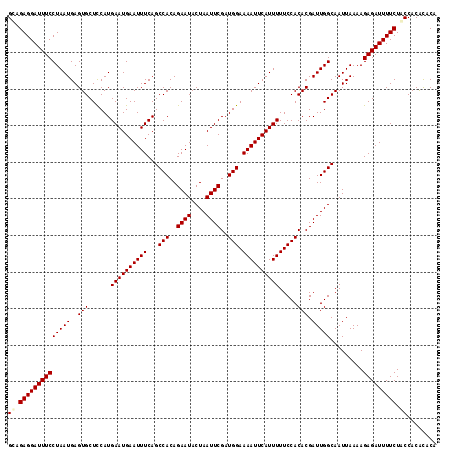
| Location | 3,405,256 – 3,405,373 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 97.44 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -26.33 |
| Energy contribution | -25.88 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3405256 117 - 22224390 UGUGCGUGGUAGAAAAUCUCUUUUAAUUGCCAAUCGUGUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGAAGCACUCAUUAGGAAAUCCUCAAC .((((.((((((..((......))..)))))).(((((......((((((((((((((.((((....)))).))))..))))))))))))))).)))).....(((....))).... ( -28.30) >DroVir_CAF1 74717 117 - 1 UGUGUGUGGUAGAAAAUCUCUUUUAAUUGCCAAUCGUGUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGGAGCACUCAUUAGGAAAUCCUCUGC ((.(((((((((..((......))..)))))..(((.((((((((.....)))))))))))........((((((..((.....))..)))))))))).))..(((....))).... ( -25.80) >DroGri_CAF1 71935 117 - 1 UGUGUGUGGUAGAAAAUCUCUUUUAAUUGCCAAUCGUGUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGGAGCACUCAUUAGGAAAUCCUCUGC ((.(((((((((..((......))..)))))..(((.((((((((.....)))))))))))........((((((..((.....))..)))))))))).))..(((....))).... ( -25.80) >DroYak_CAF1 54332 117 - 1 UGUGCGUGGUAGAAAAUCUCUUUUAAUUGCCAAUCGUGUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGAAGCACUCAUUAGGAAAUCCUCAAC .((((.((((((..((......))..)))))).(((((......((((((((((((((.((((....)))).))))..))))))))))))))).)))).....(((....))).... ( -28.30) >DroMoj_CAF1 87704 117 - 1 UGUGUGUGGCAGAAAAUCUCUUUUAAUUGCCAAUCGUGUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGGAGCACUCAUUAGGAAAUCCUCUGC ((.(((((((((..((......))..)))))..(((.((((((((.....)))))))))))........((((((..((.....))..)))))))))).))..(((....))).... ( -27.80) >DroAna_CAF1 135514 117 - 1 UGUAUGUGGUAGAAAAUCUCUUUUAAUUGCCAAUCGUGUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGAAGCACUCAUUAGGAAAUCCUCAAC ......((((((..((......))..))))))...((((.....((((((((((((((.((((....)))).))))..))))))))))......)))).....(((....))).... ( -24.00) >consensus UGUGUGUGGUAGAAAAUCUCUUUUAAUUGCCAAUCGUGUGGAAAAAUGAAUUUUCCAUCGAAUUAGUAUUCUGUGGCUGAAAUUCAUUCAUGAAGCACUCAUUAGGAAAUCCUCAAC .((((.((((((..((......))..)))))).(((((......((((((((((((((.((((....)))).))))..))))))))))))))).)))).....(((....))).... (-26.33 = -25.88 + -0.44)
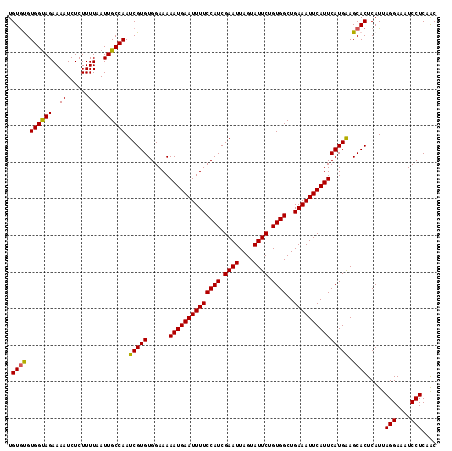
| Location | 3,405,296 – 3,405,387 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 92.12 |
| Mean single sequence MFE | -13.80 |
| Consensus MFE | -13.70 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3405296 91 + 22224390 GCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACGCACACACACACAC----ACCCU ((((..((((....))))(.(((((((.....)))))))).....)))).....................................----..... ( -13.70) >DroSec_CAF1 51705 89 + 1 GCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACGCACACA--CACAC----ACCCU ((((..((((....))))(.(((((((.....)))))))).....))))..............................--.....----..... ( -13.70) >DroSim_CAF1 53399 89 + 1 GCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACGCACACA--CACAC----ACCCU ((((..((((....))))(.(((((((.....)))))))).....))))..............................--.....----..... ( -13.70) >DroEre_CAF1 63375 93 + 1 GCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACGCACACA--CACACACGGACACA ((((..((((....))))(.(((((((.....)))))))).....))))..............................--.............. ( -13.70) >DroYak_CAF1 54372 91 + 1 GCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACGCACACA--CACACAC--AUUCA ((((..((((....))))(.(((((((.....)))))))).....))))..............................--.......--..... ( -13.70) >DroAna_CAF1 135554 90 + 1 GCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACAUACACC--CAU---CGGAAAUG ((((..((((....))))(.(((((((.....)))))))).....))))...........((((((.............--...---.)))))). ( -14.33) >consensus GCCACAGAAUACUAAUUCGAUGGAAAAUUCAUUUUUCCACACGAUUGGCAAUUAAAAGAGAUUUUCUACCACGCACACA__CACAC____ACCCU ((((..((((....))))(.(((((((.....)))))))).....)))).............................................. (-13.70 = -13.70 + 0.00)

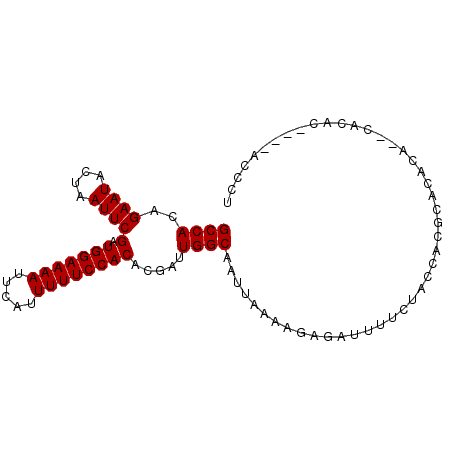
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:21 2006