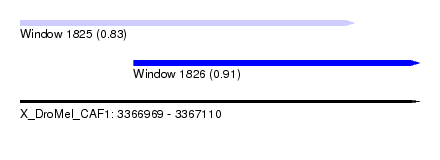
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,366,969 – 3,367,110 |
| Length | 141 |
| Max. P | 0.908036 |

| Location | 3,366,969 – 3,367,087 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -44.05 |
| Consensus MFE | -33.71 |
| Energy contribution | -35.27 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3366969 118 + 22224390 GCCGAACAAACAAACUCUCUAAGCUAGCCGAUGCAAUUCGAAGCGAGCAGAGCGCAGGAAAACCGUUCGUCACAUCUGGCGCCCAACGUGGGCGUUAGCAUGGAAGAGCUUUUCCUGU-- ................((((..(((.(((((......)))..)).))))))).(((((((((..((((.((.((((((((((((.....))))))))).))))).)))))))))))))-- ( -43.70) >DroSec_CAF1 14326 120 + 1 GACGAACGAACAAACUGGCUGAGCUAGUCGUUGCAAUUCGAAGCGAGCAGAAGGCUGGAAAACAGUUCGUCACAGCUGGCGCCCAACGUGGGCGUUAGCAGAGCAGAGCCUUUCCUGCAA (((((((.......(..(((...((..(((((.........)))))..))..)))..)......)))))))...((((((((((.....))))))))))...((((........)))).. ( -49.22) >DroSim_CAF1 14300 119 + 1 GACGAACAAACAAACUGGCUGAGCUAGUCGUUGCAAUUCGAAGCGAGCAGAAGGCAGGAAAACAGUUUGUCACAGCUGGCGCCCAACGUGGGCGG-AGCAGAGCAGAGCCUUUCCUGCAA (((((((.......(((.((..(((..(((........)))....)))...)).))).......)))))))...(((..(((((.....))))).-)))...((((........)))).. ( -39.24) >consensus GACGAACAAACAAACUGGCUGAGCUAGUCGUUGCAAUUCGAAGCGAGCAGAAGGCAGGAAAACAGUUCGUCACAGCUGGCGCCCAACGUGGGCGUUAGCAGAGCAGAGCCUUUCCUGCAA ..............(((.((..(((..(((........)))))).)))))...((((((((...((((......((((((((((.....))))))))))......)))).)))))))).. (-33.71 = -35.27 + 1.56)

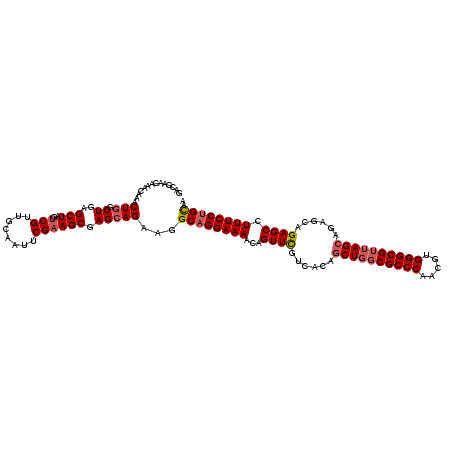
| Location | 3,367,009 – 3,367,110 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -31.01 |
| Energy contribution | -32.57 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3367009 101 + 22224390 AAGCGAGCAGAGCGCAGGAAAACCGUUCGUCACAUCUGGCGCCCAACGUGGGCGUUAGCAUGGAAGAGCUUUUCCUGU------AGCAACAUGAGAAGGCCGACAAA ..(((.((...(((((((((((..((((.((.((((((((((((.....))))))))).))))).)))))))))))))------.))..(....)...)))).)... ( -38.90) >DroSec_CAF1 14366 107 + 1 AAGCGAGCAGAAGGCUGGAAAACAGUUCGUCACAGCUGGCGCCCAACGUGGGCGUUAGCAGAGCAGAGCCUUUCCUGCAACAUGAGCAACAUGAGAAGCCCGACAAA ..((..(((((((((((.....).((((......((((((((((.....)))))))))).))))..))))))..))))..((((.....))))....))........ ( -38.50) >DroSim_CAF1 14340 106 + 1 AAGCGAGCAGAAGGCAGGAAAACAGUUUGUCACAGCUGGCGCCCAACGUGGGCGG-AGCAGAGCAGAGCCUUUCCUGCAACAUGAGCAACAUGAGAAGCCCGACAAA ..(((.((.....((((((((....(((((....(((..(((((.....))))).-)))...)))))...))))))))..((((.....))))....)).)).)... ( -33.60) >consensus AAGCGAGCAGAAGGCAGGAAAACAGUUCGUCACAGCUGGCGCCCAACGUGGGCGUUAGCAGAGCAGAGCCUUUCCUGCAACAUGAGCAACAUGAGAAGCCCGACAAA ..((...((....((((((((...((((......((((((((((.....))))))))))......)))).))))))))....)).)).................... (-31.01 = -32.57 + 1.56)

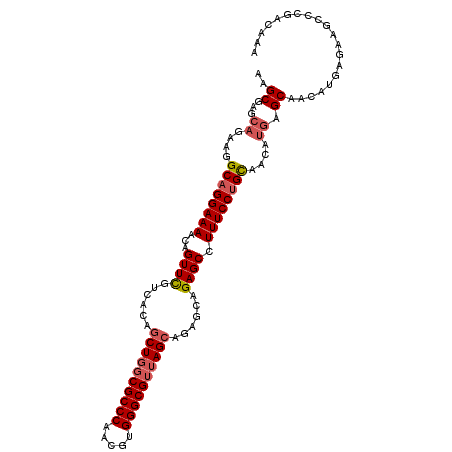
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:13 2006