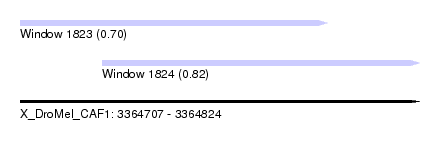
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,364,707 – 3,364,824 |
| Length | 117 |
| Max. P | 0.822318 |

| Location | 3,364,707 – 3,364,797 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -17.75 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
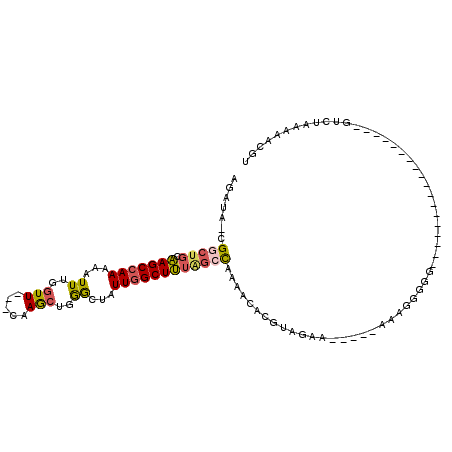
>X_DroMel_CAF1 3364707 90 + 22224390 GGCAGUCAUCACAAACUGAUGAAAAGAUA-CGGCUGCAAGCCAAAAAUUUGGUU---CAAGCUGGGCUAUUGGCUUUAGCCAAAACACGUAGAA--- (((..((((((.....)))))).......-(((((...((((((....))))))---..))))).))).(((((....)))))...........--- ( -26.70) >DroSec_CAF1 11045 90 + 1 GGCAGUCAUCUCAAACUGAUUAAAAGAUA-CGGCUGCAAGCCAAAAAUUUGGUU---CAAGCUGGGCUAUUGGCUUUAGCCAAAACACGUAGAA--- .((((((((((.............)))).-.))))))..........(((((((---.((((..(....)..)))).)))))))..........--- ( -23.42) >DroSim_CAF1 11292 90 + 1 GGCAGUCAUCACAAACUGAUGAAAAGAUA-CGGCUGCAAGCCAAAAAUUUGGUU---CAAGCUGGGCUAUUGGCUUUAGCCAAAACACGUAGAA--- (((..((((((.....)))))).......-(((((...((((((....))))))---..))))).))).(((((....)))))...........--- ( -26.70) >DroEre_CAF1 15182 90 + 1 GGCAGUCAUCACAAACUGAUGAAAAGAUA-CGACUGCAAGCCAAAAAUUUGGUU---CAAGCUGGGCUAUUGGCUUUGGCCAAAACACGUAGAA--- .((((((((((.....))))(........-)))))))..........((((((.---.((((..(....)..))))..))))))..........--- ( -23.70) >DroYak_CAF1 13986 90 + 1 GGCAGUCAUCACAAACUGAUGAAAAGAUA-CGGCUGCAAGCCAAAAAUUUGGUU---CAAGCUGGACUAUUGGCUUUGGCCAAAACACGUAGAA--- .((..((((((.....)))))).......-.(((((.(((((((.....(((((---(.....)))))))))))))))))).......))....--- ( -25.70) >DroAna_CAF1 29979 91 + 1 GCCAGUCAUC---AACUGAUGGAAAGAUAACGGCUCCGAGCCAAG--UUCCCUUACACAAAUCCAGUUAUUUGCUCUCC-UUGCACUGACAGAAAUU ..((((((((---....))))((.((((((((((.....)))..(--(......)).........)))))))..))...-....))))......... ( -16.80) >consensus GGCAGUCAUCACAAACUGAUGAAAAGAUA_CGGCUGCAAGCCAAAAAUUUGGUU___CAAGCUGGGCUAUUGGCUUUAGCCAAAACACGUAGAA___ .....(((((.......))))).........(((((.(((((((...((..(((.....)))..))...))))))))))))................ (-17.75 = -17.95 + 0.20)

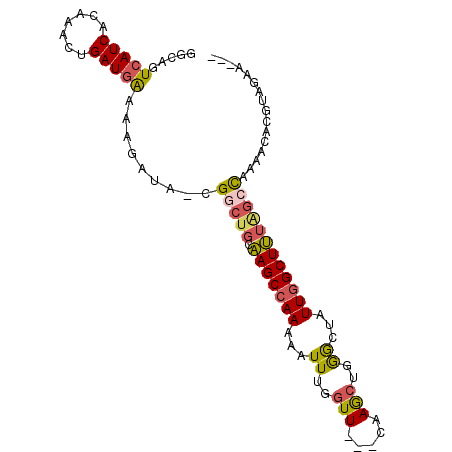
| Location | 3,364,731 – 3,364,824 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.05 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -14.55 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3364731 93 + 22224390 AGAUA-CGGCUGCAAGCCAAAAAUUUGGUU---CAAGCUGGGCUAUUGGCUUUAGCCAAAACACGUAGAA-----AAGGGGGG-----------GGGGUGUGUCUAAAAACGU (((((-((.((.(...((.....(((((((---.((((..(....)..)))).)))))))...(......-----..)))..)-----------.)).)))))))........ ( -26.80) >DroSec_CAF1 11069 86 + 1 AGAUA-CGGCUGCAAGCCAAAAAUUUGGUU---CAAGCUGGGCUAUUGGCUUUAGCCAAAACACGUAGAA-----AAAGGGGG------------------GUCUAAACUCGU .....-.(((((.(((((((.....(((((---(.....)))))))))))))))))).............-----......((------------------((....)))).. ( -22.90) >DroSim_CAF1 11316 86 + 1 AGAUA-CGGCUGCAAGCCAAAAAUUUGGUU---CAAGCUGGGCUAUUGGCUUUAGCCAAAACACGUAGAA-----AAAGGGGG------------------GUCUAAAAACGU (((..-((((.....))).....(((((((---.((((..(....)..)))).)))))))..........-----......).------------------.)))........ ( -21.50) >DroEre_CAF1 15206 86 + 1 AGAUA-CGACUGCAAGCCAAAAAUUUGGUU---CAAGCUGGGCUAUUGGCUUUGGCCAAAACACGUAGAA-----AAAGGGUG------------------GUCUAGAAACGU ....(-((.((...((((((....))))))---..))((((((..(((((....)))))..(((.(....-----...).)))------------------))))))...))) ( -21.10) >DroYak_CAF1 14010 86 + 1 AGAUA-CGGCUGCAAGCCAAAAAUUUGGUU---CAAGCUGGACUAUUGGCUUUGGCCAAAACACGUAGAA-----AAAGGGGG------------------GUCAAAAAACGU .....-(((((...((((((....))))))---..)))))((((.(((((....)))))....(.(....-----..).)..)------------------)))......... ( -21.10) >DroAna_CAF1 30000 110 + 1 AGAUAACGGCUCCGAGCCAAG--UUCCCUUACACAAAUCCAGUUAUUUGCUCUCC-UUGCACUGACAGAAAUUCCAAGGGGGUGUCUUGGCAAAGGGGUGUAUUCAUAACCAA .((..(((.((((..((((((--.((((((.......((..((((..(((.....-..))).)))).)).......))))))...))))))...)))))))..))........ ( -28.64) >consensus AGAUA_CGGCUGCAAGCCAAAAAUUUGGUU___CAAGCUGGGCUAUUGGCUUUAGCCAAAACACGUAGAA_____AAAGGGGG__________________GUCUAAAAACGU .......(((((.(((((((...((..(((.....)))..))...))))))))))))........................................................ (-14.55 = -14.72 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:11 2006