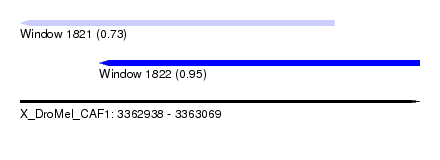
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,362,938 – 3,363,069 |
| Length | 131 |
| Max. P | 0.951029 |

| Location | 3,362,938 – 3,363,041 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -28.57 |
| Consensus MFE | -23.36 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
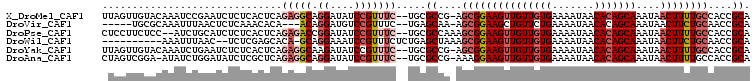
>X_DroMel_CAF1 3362938 103 - 22224390 CAGAGGCAGGAUAUCCGUUUCUGCGCCG-AGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCAAUGAGCACUGC------ACUCCCCCCCCCCUC- ..((((((((....))((((.((((..(-.(((((((((((((((.......)))))))....)))))))))..))))..))))..)))------.)))...........- ( -30.60) >DroVir_CAF1 10688 99 - 1 CA---ACAGGAUGUCCGUUUCUGAGCAA-AGCGGAAGCUGUUCUGAAAAUAACACAGCAAAUAACUUCUGCAACCGCAUUCAGCACGCU----UCUCUGCCCCC---GUC- ..---....((((...((....((((..-.(((((((.(((((((.........)))..)))).)))))))....((.....))..)))----)....))...)---)))- ( -20.20) >DroPse_CAF1 30636 108 - 1 CAGAGACCGGAUAUCCGUUUCUGCGCCAAAGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCAAUGAGCACUGAGCCUGCACCGCCCCC---UCUU (((((((.((....))))))))).((((..((((..((.(((((....))))))).(((((.....)))))..))))..)).))...(((...((...))...)---)).. ( -31.00) >DroSec_CAF1 9290 99 - 1 CAGAGGCAGGAUAUCCGUUUCUGCGCCG-AGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCAAUGAGCACUGC------ACUCCC----CCAUC- ..((((((((....))((((.((((..(-.(((((((((((((((.......)))))))....)))))))))..))))..))))..)))------.)))..----.....- ( -30.60) >DroEre_CAF1 13406 101 - 1 CAGAGGCAGGAUAUCCGUUUCUGCGCCG-AGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCAAUGAGCACUGC------ACUCCCCCC---AUCU ..((((((((....))((((.((((..(-.(((((((((((((((.......)))))))....)))))))))..))))..))))..)))------.))).....---.... ( -30.60) >DroAna_CAF1 27381 101 - 1 CAGAGGCAGGAUAUCCGUUUCUGCGCCG-AAAGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCAAUGAGCACAGA------GCUGCAUCA---GUAC .((((((.((....))))))))(((.((-((((....((((((((.......))))))))....))))))....)))..((((((....------..))).)))---.... ( -28.40) >consensus CAGAGGCAGGAUAUCCGUUUCUGCGCCG_AGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCAAUGAGCACUGC______ACUCCCCCC___AUC_ (((((((.((....))))))))).((((..((((..(.((((((....))))))).(((((.....)))))..))))..)).))........................... (-23.36 = -24.25 + 0.89)

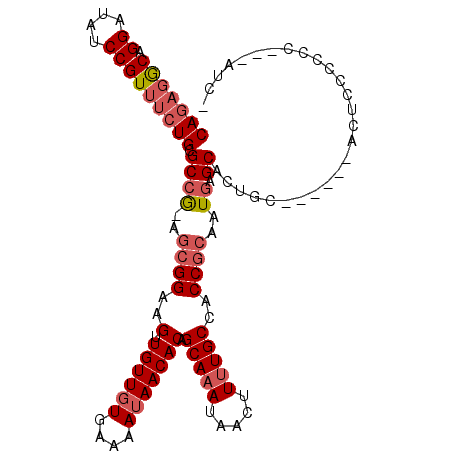
| Location | 3,362,964 – 3,363,069 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -16.99 |
| Energy contribution | -18.22 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3362964 105 - 22224390 UUAGUUGUACAAAUCCGAAUCUCUCACUCAGAGGCAGGAUAUCCGUUUC--UGCGCCG-AGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCA ..((((((.....((((...(((...(.(((((((.((....)))))))--)).)..)-))))))..((((((((.......))))))))))))))..(((....))) ( -28.30) >DroVir_CAF1 10713 97 - 1 -----UGCGCAAAUUUAACUCUCAAACACA---ACAGGAUGUCCGUUUC--UGAGCAA-AGCGGAAGCUGUUCUGAAAAUAACACAGCAAAUAACUUCUGCAACCGCA -----.(((.........(((..(((((((---......)))..)))).--.)))...-.(((((((.(((((((.........)))..)))).)))))))...))). ( -19.70) >DroPse_CAF1 30666 104 - 1 CUCCUUCUCC--AUCUGCAUCUCUCACUCAGAGACCGGAUAUCCGUUUC--UGCGCCAAAGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCA ..........--....((........(.(((((((.((....)))))))--)).).....(((((((((((((((.......)))))))....))))))))....)). ( -27.50) >DroWil_CAF1 19635 95 - 1 ----------AAAUUUAAC--UCUCGAGCACA-GCAGGAAAUCCGUUUCUCUGAGCUAAAGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUCUGCAACCGCA ----------.........--.....(((.((-(.(((((.....)))))))).)))...(((((((((((((((.......)))))))....))))))))....... ( -27.60) >DroYak_CAF1 12046 105 - 1 UUAGUUGUACAAAUCUGAAUCUCUCACUCAGAGGCAAGAUAUCCGUUUC--UGCGCCG-AGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCA ...((.((.....(((((.........)))))(((((((......((((--(((....-.)))))))((((((((.......)))))))).....))))))))).)). ( -30.10) >DroAna_CAF1 27405 104 - 1 CUAGUCGGA-AUAUCUGGAUAUCUCGCUCAGAGGCAGGAUAUCCGUUUC--UGCGCCG-AAAGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCA ......(((-(....((((((((..(((....)))..)))))))).)))--)(((.((-((((....((((((((.......))))))))....))))))....))). ( -33.80) >consensus _UAGUUGUACAAAUCUGAAUCUCUCACUCAGAGGCAGGAUAUCCGUUUC__UGCGCCA_AGCGGAAGUUGUUGUGAAAAUAACACAGCAAAUAACUUUUGCCACCGCA ..............................(((((.((....))))))).....((....(((((((((((((((.......)))))))....))))))))....)). (-16.99 = -18.22 + 1.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:09 2006