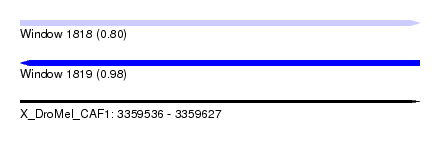
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,359,536 – 3,359,627 |
| Length | 91 |
| Max. P | 0.980081 |

| Location | 3,359,536 – 3,359,627 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -15.40 |
| Consensus MFE | -10.41 |
| Energy contribution | -10.75 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
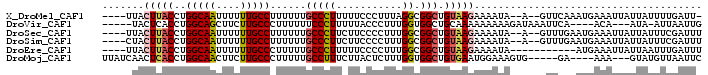
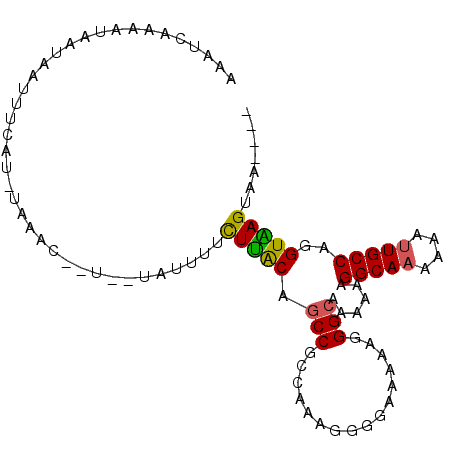
>X_DroMel_CAF1 3359536 91 + 22224390 ----UUACUUACCUGGCAAUUUUUUGCCUUUUUUGCCCUUUUUCCCUUUAGGCGGCUGUAAGAAAAUA--A--GUUCAAAUGAAAUUAUUAUUUUGAUU- ----...(((((..(((((....)))))......((((((.........))).))).)))))((((((--(--.(((....)))....)))))))....- ( -18.80) >DroVir_CAF1 5889 87 + 1 -----UACUCACCUGGCAGCUUCUUGCCCUUUUUUCCCUUUUUACCCUUUGGUGGCUGCAAAAAAAAAGAUAAAUUCA----ACA---AUA-AUUAAUUG -----.........(((((....)))))(((((((......(((((....))))).......))))))).........----...---...-........ ( -10.32) >DroSec_CAF1 5993 92 + 1 ----UUACUUACCUGGCAAUUUUUUGCCUUUUUUGCCCUUCUUCCCCUUUGGCGGCUGUAAGAAAAUA--A--GUUUGAAUGAAAUUAUUAUUUCGAUUU ----...(((((..(((((....)))))......(((((...........)).))).)))))......--.--.......((((((....)))))).... ( -16.90) >DroSim_CAF1 6022 92 + 1 ----CUACUUACCUGGCAAUUUUUUGCCUUUUUUGCCCUUCUUCCCCUUUGGCGGCUGUAAGAAAAUA--A--GUUUGAAUGAAAUUAUUAUUUCGAUUU ----...(((((..(((((....)))))......(((((...........)).))).)))))......--.--.......((((((....)))))).... ( -16.90) >DroEre_CAF1 9181 85 + 1 ----UUACUUACCUGGCAAUUUUUUGCCCUUUUUGCCCUUUUUCCCCUUUGGCGGCUGUAAGAAAAUA-----------AUGAAAUUAUUAAUUUGAUUU ----...(((((..(((((....)))))......(((((...........)).))).)))))......-----------...((((((......)))))) ( -14.80) >DroMoj_CAF1 8430 88 + 1 UUAUCAACUCACCUGGCAACUUCUUGCCCUUUUUGCCUUUCUUACUCUUUGGUGGCUGUGAAUGGAAAGUG-----GA----AAA---GUAUGUUAAUUC .....(((......(((((....)))))((((((..(((((((((.((.....))..)))...))))))..-----))----)))---)...)))..... ( -14.70) >consensus ____UUACUUACCUGGCAAUUUUUUGCCCUUUUUGCCCUUCUUCCCCUUUGGCGGCUGUAAGAAAAUA__A__GUUCA_AUGAAAUUAUUAUUUUGAUUU .......(((((..(((((....)))))......(((((...........)).))).)))))...................................... (-10.41 = -10.75 + 0.34)
| Location | 3,359,536 – 3,359,627 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -17.05 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.07 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
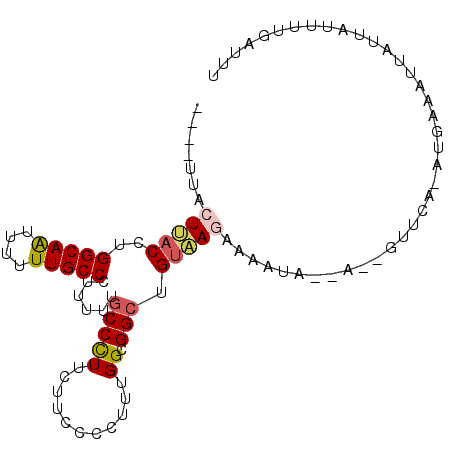
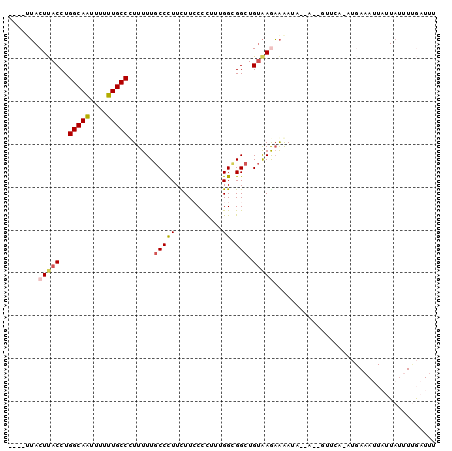
>X_DroMel_CAF1 3359536 91 - 22224390 -AAUCAAAAUAAUAAUUUCAUUUGAAC--U--UAUUUUCUUACAGCCGCCUAAAGGGAAAAAGGGCAAAAAAGGCAAAAAAUUGCCAGGUAAGUAA---- -.........(((((.(((....))).--)--))))..(((((....((((...........))))......(((((....)))))..)))))...---- ( -17.10) >DroVir_CAF1 5889 87 - 1 CAAUUAAU-UAU---UGU----UGAAUUUAUCUUUUUUUUUGCAGCCACCAAAGGGUAAAAAGGGAAAAAAGGGCAAGAAGCUGCCAGGUGAGUA----- ((((....-...---.))----)).((((((((((((((((((..((......)))))))))))))......((((......)))).))))))).----- ( -17.80) >DroSec_CAF1 5993 92 - 1 AAAUCGAAAUAAUAAUUUCAUUCAAAC--U--UAUUUUCUUACAGCCGCCAAAGGGGAAGAAGGGCAAAAAAGGCAAAAAAUUGCCAGGUAAGUAA---- .....(((((....)))))......((--(--(((((((((.(..((......)).))))))).........(((((....)))))..))))))..---- ( -17.60) >DroSim_CAF1 6022 92 - 1 AAAUCGAAAUAAUAAUUUCAUUCAAAC--U--UAUUUUCUUACAGCCGCCAAAGGGGAAGAAGGGCAAAAAAGGCAAAAAAUUGCCAGGUAAGUAG---- .....(((((....)))))......((--(--(((((((((.(..((......)).))))))).........(((((....)))))..))))))..---- ( -17.60) >DroEre_CAF1 9181 85 - 1 AAAUCAAAUUAAUAAUUUCAU-----------UAUUUUCUUACAGCCGCCAAAGGGGAAAAAGGGCAAAAAGGGCAAAAAAUUGCCAGGUAAGUAA---- .....................-----------......(((((.(((.((.....))......)))......(((((....)))))..)))))...---- ( -14.50) >DroMoj_CAF1 8430 88 - 1 GAAUUAACAUAC---UUU----UC-----CACUUUCCAUUCACAGCCACCAAAGAGUAAGAAAGGCAAAAAGGGCAAGAAGUUGCCAGGUGAGUUGAUAA ..((((((...(---(((----(.-----..(((((.((((............))))..)))))...)))))(((((....)))))......)))))).. ( -17.70) >consensus AAAUCAAAAUAAUAAUUUCAU_UAAAC__U__UAUUUUCUUACAGCCGCCAAAGGGGAAAAAGGGCAAAAAAGGCAAAAAAUUGCCAGGUAAGUAA____ ......................................(((((.(((................)))......(((((....)))))..)))))....... (-12.46 = -12.07 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:02:07 2006