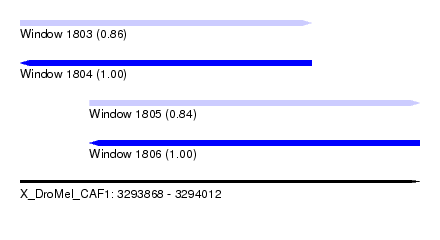
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,293,868 – 3,294,012 |
| Length | 144 |
| Max. P | 0.999996 |

| Location | 3,293,868 – 3,293,973 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.76 |
| Mean single sequence MFE | -26.43 |
| Consensus MFE | -13.54 |
| Energy contribution | -13.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3293868 105 + 22224390 GG--------UAUAAUUUGUAAAU-------UGCCAAGAAUUCUACGAGUUUGUAAUCGUCGCAACACAGAUACUCCUUUUAAUUAGAGUAUCUGCAAAGGAGACGCCUCGACCAAAUUG ((--------((((((((((((((-------(......)))).)))))).))))).(((..((....(((((((((..........)))))))))....(....)))..)))))...... ( -25.10) >DroSim_CAF1 39727 86 + 1 GG--------UAUUAUUUGUAAAU-------UG-------------------GUGAUUGGCGUAGCUCAGAUACUCCUUAUUAUUAGAGUAUCUGCAAAGGAGACGCCUCGACCAAAUUG ..--------.............(-------((-------------------((((..(((((..(((((((((((..........)))))))))....))..))))))).))))).... ( -26.30) >DroYak_CAF1 66254 114 + 1 GGUAAACUUUUGUCAUUUGUAAGUGCUCCUUGGCCUAGUGUUCGA----UU--UGAACGUCAGACCACAGGUACCCCUUAUUAUUAGAGUAUCUGAAAAGGAGACGACUCGACCAAAUUG (((......(((((....((....))(((((((.((..(((((..----..--.)))))..)).)).(((((((..((.......)).)))))))..))))))))))....)))...... ( -27.90) >consensus GG________UAUAAUUUGUAAAU_______UGCC_AG__UUC_A____UU_GUGAUCGUCGCAACACAGAUACUCCUUAUUAUUAGAGUAUCUGCAAAGGAGACGCCUCGACCAAAUUG ..........................................................(((......(((((((((..........)))))))))....(....))))............ (-13.54 = -13.77 + 0.23)

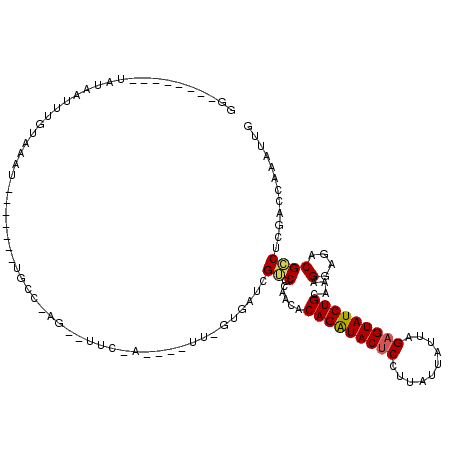
| Location | 3,293,868 – 3,293,973 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.76 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -16.29 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.56 |
| SVM decision value | 6.09 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3293868 105 - 22224390 CAAUUUGGUCGAGGCGUCUCCUUUGCAGAUACUCUAAUUAAAAGGAGUAUCUGUGUUGCGACGAUUACAAACUCGUAGAAUUCUUGGCA-------AUUUACAAAUUAUA--------CC .(((((((((((((((((..(..((((((((((((........))))))))))))..).)))).((((......))))...))))))).-------.....))))))...--------.. ( -29.90) >DroSim_CAF1 39727 86 - 1 CAAUUUGGUCGAGGCGUCUCCUUUGCAGAUACUCUAAUAAUAAGGAGUAUCUGAGCUACGCCAAUCAC-------------------CA-------AUUUACAAAUAAUA--------CC ....(((((.(((((((........((((((((((........))))))))))....)))))..))))-------------------))-------).............--------.. ( -25.90) >DroYak_CAF1 66254 114 - 1 CAAUUUGGUCGAGUCGUCUCCUUUUCAGAUACUCUAAUAAUAAGGGGUACCUGUGGUCUGACGUUCA--AA----UCGAACACUAGGCCAAGGAGCACUUACAAAUGACAAAAGUUUACC ..(((((...((((.(.((((((..(((.((((((........)))))).))).((((((..((((.--..----..))))..))))))))))))))))).))))).............. ( -32.00) >consensus CAAUUUGGUCGAGGCGUCUCCUUUGCAGAUACUCUAAUAAUAAGGAGUAUCUGUGCUACGACGAUCAC_AA____U_GAA__CU_GGCA_______AUUUACAAAUAAUA________CC .......((((..((..........((((((((((........)))))))))).))..)))).......................................................... (-16.29 = -15.97 + -0.33)

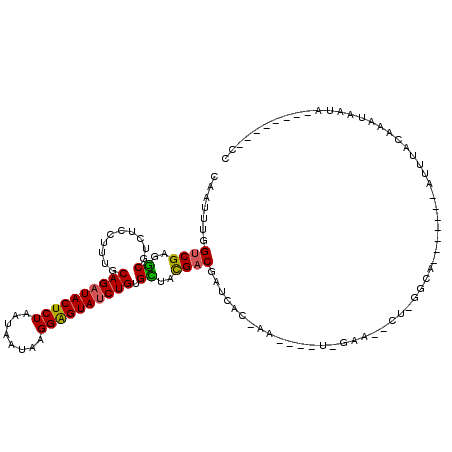
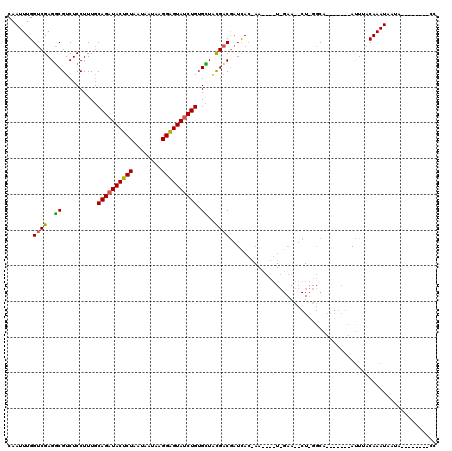
| Location | 3,293,893 – 3,294,012 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -21.99 |
| Energy contribution | -25.77 |
| Covariance contribution | 3.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3293893 119 + 22224390 UUCUACGAGUUUGUAAUCGUCGCAACACAGAUACUCCUUUUAAUUAGAGUAUCUGCAAAGGAGACGCCUCGACCAAAUUGAUCGCGGCAGUCCACAAAAAAAAAAAAGUGCGUGAGCGA .........(((((....(((.(....(((((((((..........))))))))).....).)))(((.(((.(.....).))).))).....)))))..........(((....))). ( -31.10) >DroSim_CAF1 39745 100 + 1 ------------GUGAUUGGCGUAGCUCAGAUACUCCUUAUUAUUAGAGUAUCUGCAAAGGAGACGCCUCGACCAAAUUGAUCGGGGCAGUCCA-------AAAAAAGUGCGUGAGCGA ------------....(((((((..(((((((((((..........)))))))))....))..))(((((((.(.....).))))))).).)))-------)......(((....))). ( -33.60) >DroYak_CAF1 66294 106 + 1 UUCGA----UU--UGAACGUCAGACCACAGGUACCCCUUAUUAUUAGAGUAUCUGAAAAGGAGACGACUCGACCAAAUUGAUCGGGGCAGUCCA-------AAAAAAGUGCGUGAGCGA .((((----((--((...(((((....(((((((..((.......)).)))))))....(....)..)).)))))))))))..(((....))).-------.......(((....))). ( -24.80) >consensus UUC_A____UU_GUGAUCGUCGCAACACAGAUACUCCUUAUUAUUAGAGUAUCUGCAAAGGAGACGCCUCGACCAAAUUGAUCGGGGCAGUCCA_______AAAAAAGUGCGUGAGCGA ...........................(((((((((..........)))))))))....(((...(((((((.(.....).)))))))..)))...............(((....))). (-21.99 = -25.77 + 3.78)

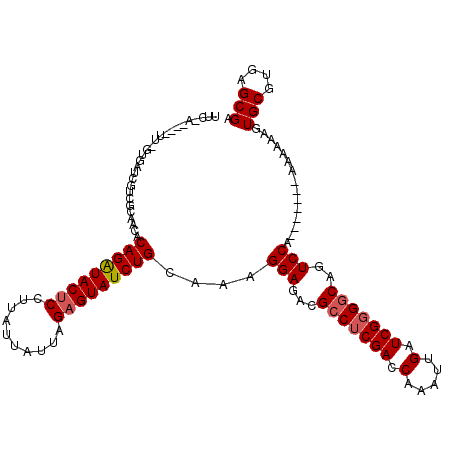
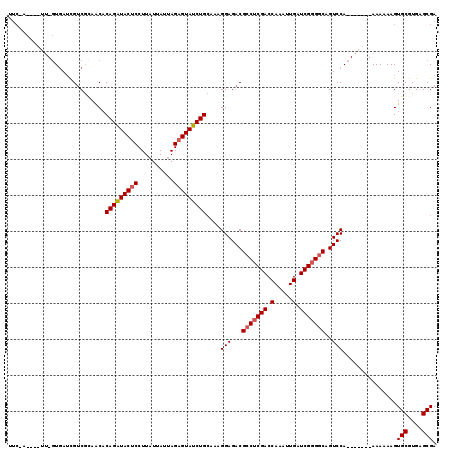
| Location | 3,293,893 – 3,294,012 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.84 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -26.22 |
| Energy contribution | -26.67 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3293893 119 - 22224390 UCGCUCACGCACUUUUUUUUUUUUGUGGACUGCCGCGAUCAAUUUGGUCGAGGCGUCUCCUUUGCAGAUACUCUAAUUAAAAGGAGUAUCUGUGUUGCGACGAUUACAAACUCGUAGAA ....((.((((.............(..(((.(((.(((((.....))))).))))))..)...(((((((((((........)))))))))))..))))((((........)))).)). ( -39.80) >DroSim_CAF1 39745 100 - 1 UCGCUCACGCACUUUUUU-------UGGACUGCCCCGAUCAAUUUGGUCGAGGCGUCUCCUUUGCAGAUACUCUAAUAAUAAGGAGUAUCUGAGCUACGCCAAUCAC------------ ..((((..(((.......-------.((((.(((.(((((.....))))).)))))))....)))(((((((((........)))))))))))))............------------ ( -34.60) >DroYak_CAF1 66294 106 - 1 UCGCUCACGCACUUUUUU-------UGGACUGCCCCGAUCAAUUUGGUCGAGUCGUCUCCUUUUCAGAUACUCUAAUAAUAAGGGGUACCUGUGGUCUGACGUUCA--AA----UCGAA ((((....))......((-------(((((.....(((((.....))))).((((...((....(((.((((((........)))))).))).))..)))))))))--))----..)). ( -26.90) >consensus UCGCUCACGCACUUUUUU_______UGGACUGCCCCGAUCAAUUUGGUCGAGGCGUCUCCUUUGCAGAUACUCUAAUAAUAAGGAGUAUCUGUGCUACGACGAUCAC_AA____U_GAA ..((....))................((((.(((.(((((.....))))).)))))))......((((((((((........))))))))))........................... (-26.22 = -26.67 + 0.45)

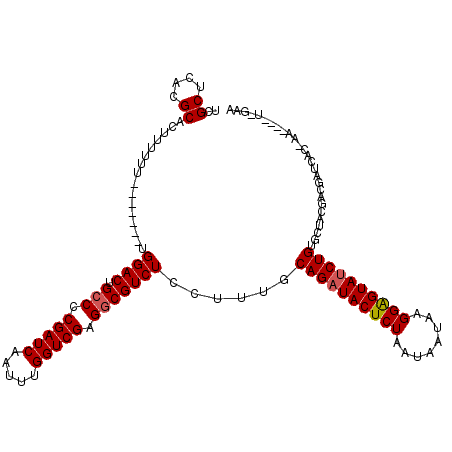
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:54 2006