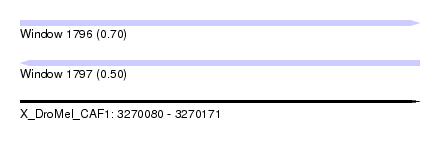
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,270,080 – 3,270,171 |
| Length | 91 |
| Max. P | 0.702581 |

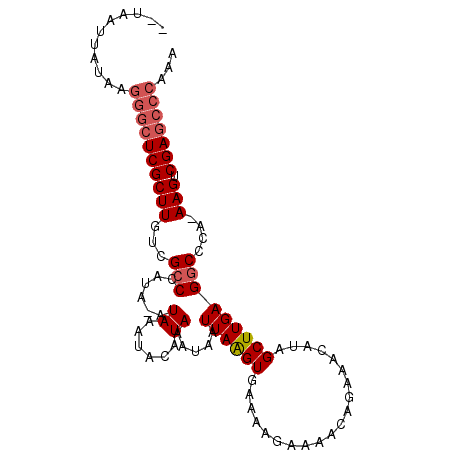
| Location | 3,270,080 – 3,270,171 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 90.47 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -14.41 |
| Energy contribution | -15.47 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3270080 91 + 22224390 --UAAUUAUAAGGGCUCGCUUGUCGCCCAUAAAUAAAUACAUAAAUAAUUAAGUGAAAAGAAAACCGAAACAUAGCUUGAGGCCCG-AAGUCGA------- --.........(((((((((((...........................)))))))..............((.....))..)))).-.......------- ( -11.93) >DroSim_CAF1 21161 97 + 1 --UAAUUAUAAGGGCUCGCUUGUCGCCCAUA-AUAAAUACAUAAAUAAUUAAGUGAAAAGAAAACAGAAACAUAGCUUGAGGCCCA-AAGUCGAGCCCAAA --.........((((((((((...(((....-................((((((....................)))))))))...-))).)))))))... ( -21.70) >DroEre_CAF1 48260 100 + 1 UGUAAUUAUAAGGGCUCGCUUGUCGCCUAUA-AUAAAUACAUAAAUAAUUAGGUGAAAAGAAAACAGAAACAUAGCUUGAGGCGCAAAAGUCGAGCCCAAA ...........((((((((((.(((((((..-.................)))))))..................((.....))....))).)))))))... ( -26.21) >DroYak_CAF1 47336 99 + 1 UGUAAUUAUAAGGGCUCGCUUGUCGGCUAUA-AUAAAUACAUAAAUAAUUAAGUGAAAAAAAAACAGAAACAUAGCUUGAGGCCCA-AAGUCGAGCCCAAA ...........((((((((((...((((...-................((((((....................))))))))))..-))).)))))))... ( -23.05) >consensus __UAAUUAUAAGGGCUCGCUUGUCGCCCAUA_AUAAAUACAUAAAUAAUUAAGUGAAAAGAAAACAGAAACAUAGCUUGAGGCCCA_AAGUCGAGCCCAAA ...........((((((((((...(((......((......)).....((((((....................)))))))))....))).)))))))... (-14.41 = -15.47 + 1.06)

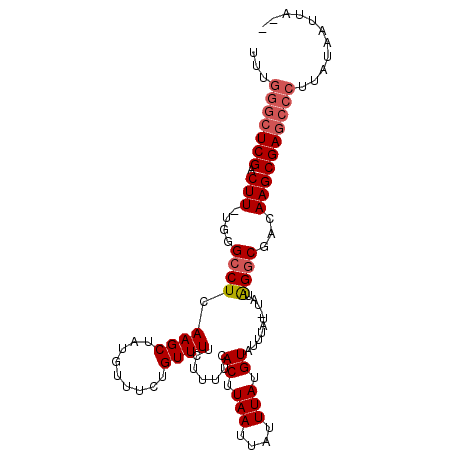
| Location | 3,270,080 – 3,270,171 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 90.47 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -14.45 |
| Energy contribution | -15.45 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3270080 91 - 22224390 -------UCGACUU-CGGGCCUCAAGCUAUGUUUCGGUUUUCUUUUCACUUAAUUAUUUAUGUAUUUAUUUAUGGGCGACAAGCGAGCCCUUAUAAUUA-- -------.......-.((((.((..(((.((((((.((.........((.(((....))).))........)).)).))))))))))))).........-- ( -12.73) >DroSim_CAF1 21161 97 - 1 UUUGGGCUCGACUU-UGGGCCUCAAGCUAUGUUUCUGUUUUCUUUUCACUUAAUUAUUUAUGUAUUUAU-UAUGGGCGACAAGCGAGCCCUUAUAAUUA-- ...(((((((.(((-.(.((((.((((.........)))).......((.(((....))).))......-...))))..))))))))))).........-- ( -21.00) >DroEre_CAF1 48260 100 - 1 UUUGGGCUCGACUUUUGCGCCUCAAGCUAUGUUUCUGUUUUCUUUUCACCUAAUUAUUUAUGUAUUUAU-UAUAGGCGACAAGCGAGCCCUUAUAAUUACA ...(((((((.(((...(((((.((((.........)))).......((.(((....))).))......-...)))))..))))))))))........... ( -24.00) >DroYak_CAF1 47336 99 - 1 UUUGGGCUCGACUU-UGGGCCUCAAGCUAUGUUUCUGUUUUUUUUUCACUUAAUUAUUUAUGUAUUUAU-UAUAGCCGACAAGCGAGCCCUUAUAAUUACA ...(((((((.(((-(.(((...((((.........))))....................((((.....-))))))).).))))))))))........... ( -21.20) >consensus UUUGGGCUCGACUU_UGGGCCUCAAGCUAUGUUUCUGUUUUCUUUUCACUUAAUUAUUUAUGUAUUUAU_UAUAGGCGACAAGCGAGCCCUUAUAAUUA__ ...(((((((.(((....((((.((((.........)))).......((.(((....))).))..........))))...))))))))))........... (-14.45 = -15.45 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:46 2006