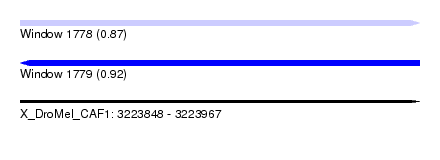
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,223,848 – 3,223,967 |
| Length | 119 |
| Max. P | 0.917153 |

| Location | 3,223,848 – 3,223,967 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -30.74 |
| Energy contribution | -30.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
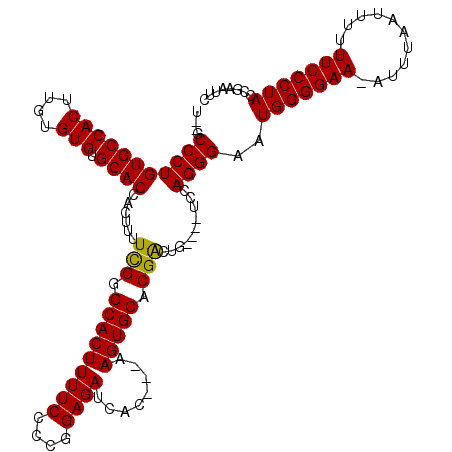
>X_DroMel_CAF1 3223848 119 + 22224390 UGUGCCCUGUGCCACUUGUGUGUGCACCACUUUUCGGGCACUUUUCCCCAGAGAUCACAACAAGUGCACGAGUGUCCGUCCAGGGAAUGGGGAA-AUUUGGUAUUUUCCCUAACGAAUUC ....(((((..((((((((((((((.((.......)))))).((((....))))...........)))))))))...)..)))))..(((((((-(........))))))))........ ( -39.80) >DroSec_CAF1 32771 110 + 1 U--GCCCUGUGCCACUUGUGUGUGCACCACUUUUCGGGCACUUUUCCCCGGAGAUCAC---AAGUGCACGACUG----UCCAGGGAAUGGGGAA-UUUUAAAUUUUUCCCUACCGAAUUC .--.((((((((.(((((((.((((.((.......)))))).((((....)))).)))---))))))))(....----..)))))..(((((((-..........)))))))........ ( -36.50) >DroYak_CAF1 38162 111 + 1 U--GCCCUGUGCCACUUGUGUGUGCACCACUUUUUGGGCACUUUUCCCCGGAGAUCAC---AAGUGCACGGCUG----UCCAGGGAAUGGGGAAAAUUUACUUUUUUCCCUACCGAAUUC .--.((((((((.(((((((.((((.(((.....))))))).((((....)))).)))---))))))))((...----.))))))..(((((((((.......)))))))))........ ( -42.10) >consensus U__GCCCUGUGCCACUUGUGUGUGCACCACUUUUCGGGCACUUUUCCCCGGAGAUCAC___AAGUGCACGACUG____UCCAGGGAAUGGGGAA_AUUUAAUUUUUUCCCUACCGAAUUC ....(((((((((((....))).))))......(((.(((((((((....))))........))))).)))..........))))..(((((((...........)))))))........ (-30.74 = -30.30 + -0.44)

| Location | 3,223,848 – 3,223,967 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -32.65 |
| Energy contribution | -32.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
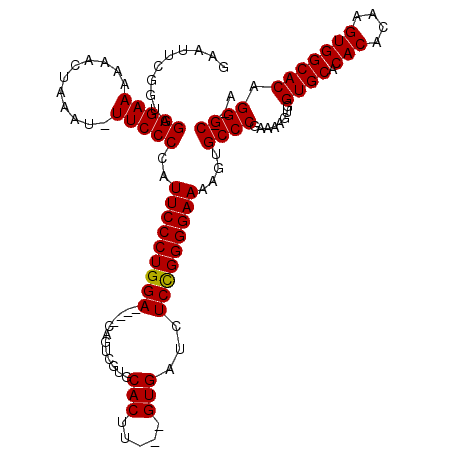
>X_DroMel_CAF1 3223848 119 - 22224390 GAAUUCGUUAGGGAAAAUACCAAAU-UUCCCCAUUCCCUGGACGGACACUCGUGCACUUGUUGUGAUCUCUGGGGAAAAGUGCCCGAAAAGUGGUGCACACACAAGUGGCACAGGGCACA ..........((((((........)-)))))..(((((..((.((.(((....((....)).))).))))..)))))..((((((........((((.(((....))))))).)))))). ( -43.10) >DroSec_CAF1 32771 110 - 1 GAAUUCGGUAGGGAAAAAUUUAAAA-UUCCCCAUUCCCUGGA----CAGUCGUGCACUU---GUGAUCUCCGGGGAAAAGUGCCCGAAAAGUGGUGCACACACAAGUGGCACAGGGC--A ..........(((((..........-)))))..(((((((((----..((((.......---.)))).)))))))))...(((((........((((.(((....))))))).))))--) ( -36.70) >DroYak_CAF1 38162 111 - 1 GAAUUCGGUAGGGAAAAAAGUAAAUUUUCCCCAUUCCCUGGA----CAGCCGUGCACUU---GUGAUCUCCGGGGAAAAGUGCCCAAAAAGUGGUGCACACACAAGUGGCACAGGGC--A ..........(((((((.......)))))))....((((((.----...))((((((((---(((.(((....)))...(((((((.....))).)))).))))))).)))))))).--. ( -39.90) >consensus GAAUUCGGUAGGGAAAAAACUAAAU_UUCCCCAUUCCCUGGA____CAGUCGUGCACUU___GUGAUCUCCGGGGAAAAGUGCCCGAAAAGUGGUGCACACACAAGUGGCACAGGGC__A ..........(((((...........)))))..(((((((((............(((.....)))...)))))))))....((((........((((.(((....))))))).))))... (-32.65 = -32.43 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:29 2006