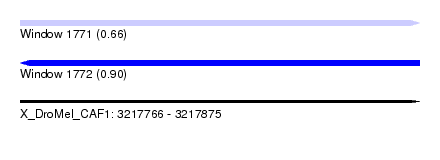
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 3,217,766 – 3,217,875 |
| Length | 109 |
| Max. P | 0.903189 |

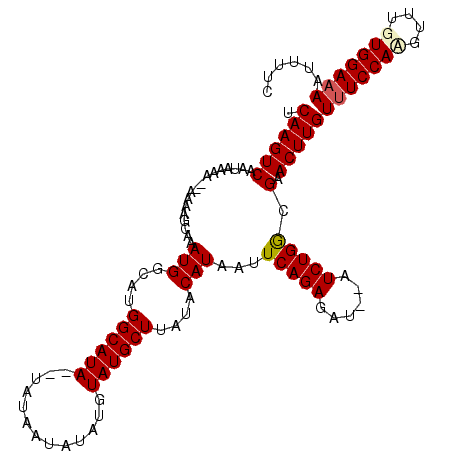
| Location | 3,217,766 – 3,217,875 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3217766 109 + 22224390 GAAAAUUUUCCACAAACAUGGAAACAAGUUCCUCAGAUAUAUCUCUGAAUUAUGUAUAAGCAUAC--AUAUUAUA--UAUGCCAUGCCAUUUGCCUUU-UUUUUAUUGACUUGA ......((((((......))))))((((((..(((((......))))).....(((...((((.(--((((...)--))))..))))....)))....-........)))))). ( -19.30) >DroSec_CAF1 26852 110 + 1 GAAAAUUUUCCACAAACUUGGAAACAAGUUCCCCAGAUAUAUCUCUGAAUUAUGUAUAAGCAUACACAUAUUAUA--UAUGCCAUGCCAUUUGCCUUU--UUUUAUUGACUUGA ......((((((......))))))((((((...((((......))))......(((...((((...(((((...)--))))..))))....)))....--.......)))))). ( -17.50) >DroEre_CAF1 28630 108 + 1 GAAA--UGUCCACAAACGUGGAAACAAGUUCGCCAGAU--AUCUCUGAAUUAUGUAUAAGCAUACAUGUAUUAUA--UAUGCCAUGCCAUUUGCUUUUUUUUUUAUUGACUUGA ....--..(((((....)))))..((((((...((((.--...))))......(((...((((.((((((...))--))))..))))....))).............)))))). ( -21.30) >DroYak_CAF1 32023 110 + 1 GAAAAUUUUCCACAAACUUGGAAACAAGUUCGCCAGAU--AUCUCUGAAUUAUGUAUAAGCAUACAUAUUAUAUAUGUAUGCCAUGGCAUUUGAUUUU--UUUUAUCGACUUGA ......((((((......))))))((((((.....(((--(.....(((((((((....((((((((((...))))))))))....))))..))))).--...)))))))))). ( -26.30) >consensus GAAAAUUUUCCACAAACUUGGAAACAAGUUCCCCAGAU__AUCUCUGAAUUAUGUAUAAGCAUACAUAUAUUAUA__UAUGCCAUGCCAUUUGCCUUU__UUUUAUUGACUUGA ......((((((......))))))((((((...((((......))))......((((..(((((.............))))).))))....................)))))). (-15.02 = -15.52 + 0.50)

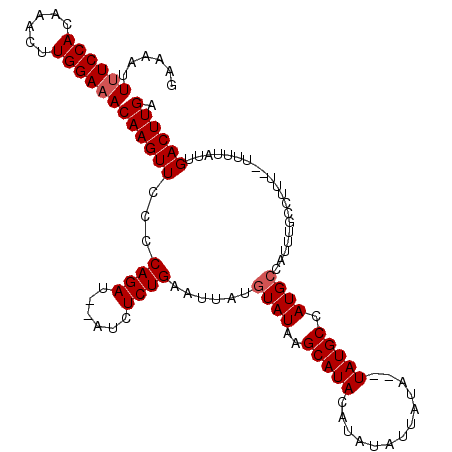
| Location | 3,217,766 – 3,217,875 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -18.75 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 3217766 109 - 22224390 UCAAGUCAAUAAAAA-AAAGGCAAAUGGCAUGGCAUA--UAUAAUAU--GUAUGCUUAUACAUAAUUCAGAGAUAUAUCUGAGGAACUUGUUUCCAUGUUUGUGGAAAAUUUUC .((((((........-........(((....((((((--(((...))--)))))))....)))..((((((......))))))).)))))(((((((....)))))))...... ( -26.00) >DroSec_CAF1 26852 110 - 1 UCAAGUCAAUAAAA--AAAGGCAAAUGGCAUGGCAUA--UAUAAUAUGUGUAUGCUUAUACAUAAUUCAGAGAUAUAUCUGGGGAACUUGUUUCCAAGUUUGUGGAAAAUUUUC .((((((.......--........(((....((((((--(((.....)))))))))....)))..((((((......))))))).)))))((((((......))))))...... ( -24.80) >DroEre_CAF1 28630 108 - 1 UCAAGUCAAUAAAAAAAAAAGCAAAUGGCAUGGCAUA--UAUAAUACAUGUAUGCUUAUACAUAAUUCAGAGAU--AUCUGGCGAACUUGUUUCCACGUUUGUGGACA--UUUC .((((((.................(((....((((((--(((.....)))))))))....)))...(((((...--.))))).).)))))..(((((....)))))..--.... ( -24.20) >DroYak_CAF1 32023 110 - 1 UCAAGUCGAUAAAA--AAAAUCAAAUGCCAUGGCAUACAUAUAUAAUAUGUAUGCUUAUACAUAAUUCAGAGAU--AUCUGGCGAACUUGUUUCCAAGUUUGUGGAAAAUUUUC .(((((((......--........(((....(((((((((((...)))))))))))....)))...(((((...--.))))))).)))))((((((......))))))...... ( -27.50) >consensus UCAAGUCAAUAAAA__AAAAGCAAAUGGCAUGGCAUA__UAUAAUAUAUGUAUGCUUAUACAUAAUUCAGAGAU__AUCUGGCGAACUUGUUUCCAAGUUUGUGGAAAAUUUUC .((((((.................(((....((((((.............))))))....)))...(((((......))))).).)))))(((((((....)))))))...... (-18.75 = -19.25 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:01:23 2006